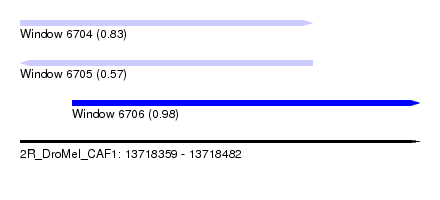
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,718,359 – 13,718,482 |
| Length | 123 |
| Max. P | 0.976573 |

| Location | 13,718,359 – 13,718,449 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 91.42 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.44 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13718359 90 + 20766785 GUUGGGGCAAAUGGAAAAGCUGCAACUGCAUAUGCAACGCCAGCA-ACUGCAACUUCAGAAUUUGCAGCCACACAACGGCAACGGCAACUG ((((.(((..........))).))))(((....)))..(((.(((-((((......)))...)))).(((.......)))...)))..... ( -24.20) >DroSec_CAF1 19518 84 + 1 GUUGGGGCAAAUGGAAAAGCUGCAAC------UGCAACGCCAGCG-ACUGCAACUUCAGAAUUUGCAGCCACACAACGGCAACGGCAACUG ((((..((((((....(((.((((..------(((.......)))-..)))).)))....)))))).(((.......)))..))))..... ( -22.40) >DroSim_CAF1 21867 90 + 1 GUUGGGGCAAAUGGAAAAGCUGCAACUGCAACUGCAACGCAAGCA-ACUGCAACUUCAGAAUUUGCAGCCACACAACGGCAACGGCAACUG ((((..((...((.....((((((((((....((((.(....)..-..))))....)))...)))))))....))...))..))))..... ( -24.50) >DroEre_CAF1 19200 84 + 1 GUUGGGGCAAAUGGAAAAGCUGCAAC------UGCAACACCAGCA-ACUGCAACUUCAGCAUCUGCAACCACACAACGGCAACGGCAACUG (((((((((.(((...(((.((((..------(((.......)))-..)))).)))...))).)))..))......((....)).)))).. ( -21.40) >DroYak_CAF1 20278 85 + 1 GUUGGGGCAAAUGGAAAAGCUGCAAC------UGCAACACCAGCAAACUGCAUCUUCAGAAGUUGCAGCCACACAACGGCAACGGCAACUG ((((..((...((.....((((((((------(((.......))...(((......))).)))))))))....))...))..))))..... ( -25.60) >consensus GUUGGGGCAAAUGGAAAAGCUGCAAC______UGCAACGCCAGCA_ACUGCAACUUCAGAAUUUGCAGCCACACAACGGCAACGGCAACUG ((((..((...((.....(((((((.......(((.......)))..(((......)))...)))))))....))...))..))))..... (-19.20 = -19.44 + 0.24)

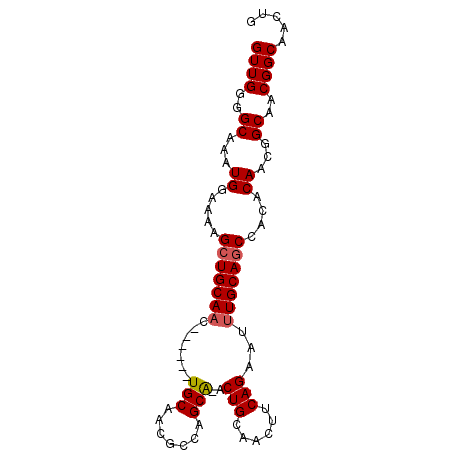
| Location | 13,718,359 – 13,718,449 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 91.42 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -21.36 |
| Energy contribution | -21.64 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13718359 90 - 20766785 CAGUUGCCGUUGCCGUUGUGUGGCUGCAAAUUCUGAAGUUGCAGU-UGCUGGCGUUGCAUAUGCAGUUGCAGCUUUUCCAUUUGCCCCAAC ..((((.....((((.....)))).((((((...(((((((((..-(((..((...))....)))..)))))))))...))))))..)))) ( -29.20) >DroSec_CAF1 19518 84 - 1 CAGUUGCCGUUGCCGUUGUGUGGCUGCAAAUUCUGAAGUUGCAGU-CGCUGGCGUUGCA------GUUGCAGCUUUUCCAUUUGCCCCAAC .((((((..(((((((((.((((((((((.........)))))))-))))))))..)))------)..))))))................. ( -29.00) >DroSim_CAF1 21867 90 - 1 CAGUUGCCGUUGCCGUUGUGUGGCUGCAAAUUCUGAAGUUGCAGU-UGCUUGCGUUGCAGUUGCAGUUGCAGCUUUUCCAUUUGCCCCAAC ..((((.....((((.....)))).((((((...(((((((((..-(((..((......)).)))..)))))))))...))))))..)))) ( -30.00) >DroEre_CAF1 19200 84 - 1 CAGUUGCCGUUGCCGUUGUGUGGUUGCAGAUGCUGAAGUUGCAGU-UGCUGGUGUUGCA------GUUGCAGCUUUUCCAUUUGCCCCAAC (((..((....))..)))..(((..(((((((..(((((((((..-(((.......)))------..)))))))))..))))))).))).. ( -31.20) >DroYak_CAF1 20278 85 - 1 CAGUUGCCGUUGCCGUUGUGUGGCUGCAACUUCUGAAGAUGCAGUUUGCUGGUGUUGCA------GUUGCAGCUUUUCCAUUUGCCCCAAC .((((((.((..(......)..)).)))))).........((((.....(((.(((((.------...)))))....))).))))...... ( -22.50) >consensus CAGUUGCCGUUGCCGUUGUGUGGCUGCAAAUUCUGAAGUUGCAGU_UGCUGGCGUUGCA______GUUGCAGCUUUUCCAUUUGCCCCAAC ..((((.....((((.....)))).((((((...((((((((((..(((.......))).......))))))))))...))))))..)))) (-21.36 = -21.64 + 0.28)

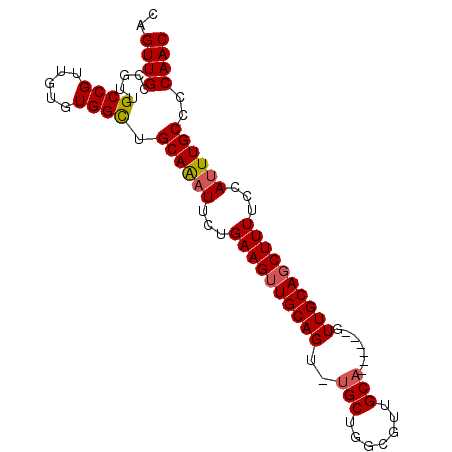
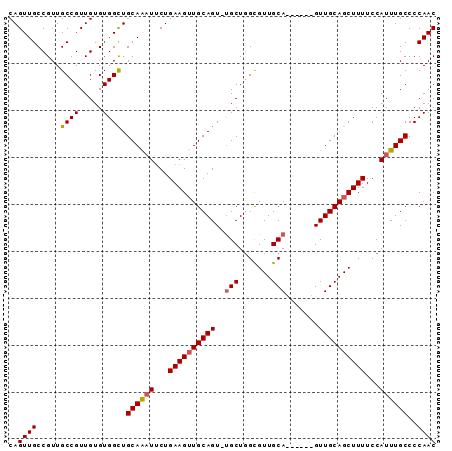
| Location | 13,718,375 – 13,718,482 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 91.71 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -27.38 |
| Energy contribution | -28.12 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13718375 107 + 20766785 AAGCUGCAACUGCAUAUGCAACGCCAGCA-ACUGCAACUUCAGAAUUUGCAGCCACACAACGGCAACGGCAACUGGCGAUAAGCAGCUCGGGAAAACUGAGAAGCCAA ..(((((...(((....))).((((((..-.((((((.........))))))........((....))....))))))....)))))((((.....))))........ ( -34.90) >DroSec_CAF1 19534 101 + 1 AAGCUGCAAC------UGCAACGCCAGCG-ACUGCAACUUCAGAAUUUGCAGCCACACAACGGCAACGGCAACUGGCGAUAAGCAGCUCGGGAAAACUGAGAAACCGA ..(((((...------.))..((((((.(-.((((((.........)))))).)......((....))....))))))...)))..(((((.....)))))....... ( -31.40) >DroSim_CAF1 21883 107 + 1 AAGCUGCAACUGCAACUGCAACGCAAGCA-ACUGCAACUUCAGAAUUUGCAGCCACACAACGGCAACGGCAACUGGCGAUAAGCAGCUCGGGAAAACUGAGAAGCCGA ..((((((((((....((((.(....)..-..))))....)))...))))))).......((((..((.(....).))........(((((.....)))))..)))). ( -32.10) >DroYak_CAF1 20294 102 + 1 AAGCUGCAAC------UGCAACACCAGCAAACUGCAUCUUCAGAAGUUGCAGCCACACAACGGCAACGGCAACUGGCGAUAAGCAGCUCGGGAAAACUGAGAAGCCGC ..((((((((------(((.......))...(((......))).))))))))).......((((..((.(....).))........(((((.....)))))..)))). ( -32.50) >consensus AAGCUGCAAC______UGCAACGCCAGCA_ACUGCAACUUCAGAAUUUGCAGCCACACAACGGCAACGGCAACUGGCGAUAAGCAGCUCGGGAAAACUGAGAAGCCGA ..((((((........)))..((((((....((((((.........))))))........((....))....))))))...)))..(((((.....)))))....... (-27.38 = -28.12 + 0.75)

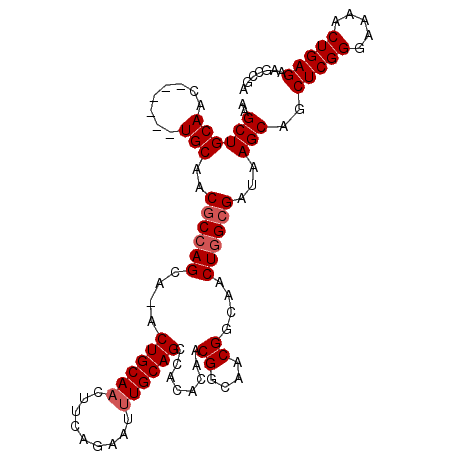
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:25 2006