
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,712,539 – 13,712,632 |
| Length | 93 |
| Max. P | 0.654878 |

| Location | 13,712,539 – 13,712,632 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.64 |
| Mean single sequence MFE | -19.85 |
| Consensus MFE | -13.86 |
| Energy contribution | -14.75 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
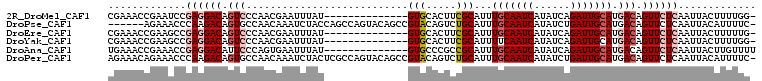
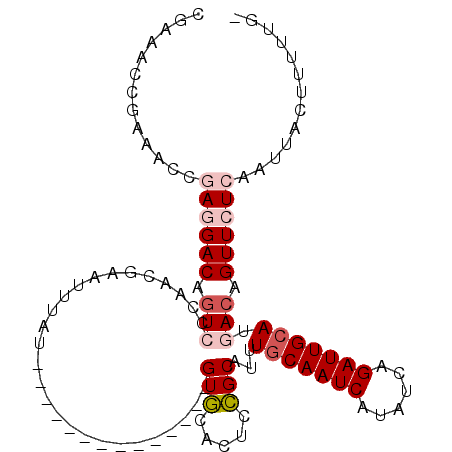
>2R_DroMel_CAF1 13712539 93 + 20766785 CGAAACCGAAUCCGAGGACAGUCCCAACGAAUUUAU--------------GUGCACUUCGCAUUUGCAAUCAUAUCAGAUUGCAUGACAGUUCUCAAUUACUUUUGG- .....(((((...((((((.(((.............--------------((((.....)))).(((((((......))))))).))).)))))).......)))))- ( -23.10) >DroPse_CAF1 9757 101 + 1 ------AGAAACCCAAGACAGUGCCAACAAAUCUACCAGCCAGUACAGCCGUACAGUCUGCAUUUGCAAUCAUAUCUGAUUGCAUGACAGUUCUCAAUUACAUUUUC- ------((((.....((((.((....))..............((((....)))).)))).....((((((((....))))))))......)))).............- ( -17.90) >DroEre_CAF1 13191 93 + 1 CGAAACCGAAGCCGAGGACAGUCCCAACGAAUUUAU--------------GUGCACUUCGCAUUUGCAAUCAUAUCAGAUUGCAUGACAGUUCUCAAUUACUUUUUG- .............((((((.(((.............--------------((((.....)))).(((((((......))))))).))).))))))............- ( -22.20) >DroYak_CAF1 14349 93 + 1 CGAAACCGAAGCCGAGGACAGUCCCAACGAAUUUAU--------------GUGCACUUCGCAUUUUCAAUCAUAUCAGAUUGCAUGACAGUUCUCAAUUACUUUUGG- .....((((((..((((((.(((.............--------------((((.....))))...(((((......)))))...))).))))))......))))))- ( -17.90) >DroAna_CAF1 11963 94 + 1 UGAAACCGAAACCGAGGACAUUCCCAGUGAAUUUAU--------------GUGCCCGCCGCAUUUGCAAUCAUAUCAGAUUGCAUGACAGUUCUCAAUUACUUGUUUU .............((((((.................--------------((((.....)))).(((((((......))))))).....))))))............. ( -19.20) >DroPer_CAF1 9639 107 + 1 AGAAACAGAAACCCAAGACAGUGCCAACAAAUCUACUCGCCAGUACAGCCGUACAGUCUGCAUUUGCAAUCAUAUCUGAUUGCAUGACAGUUCUCAAUUACAUUUUC- .(((((.........((((.((....))..............((((....)))).)))).....((((((((....)))))))).....))).))............- ( -18.80) >consensus CGAAACCGAAACCGAGGACAGUCCCAACGAAUUUAU______________GUGCACUCCGCAUUUGCAAUCAUAUCAGAUUGCAUGACAGUUCUCAAUUACUUUUUG_ .............((((((.(((...........................(((.....)))...(((((((......))))))).))).))))))............. (-13.86 = -14.75 + 0.89)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:23 2006