| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,695,014 – 13,695,124 |
| Length | 110 |
| Max. P | 1.000000 |

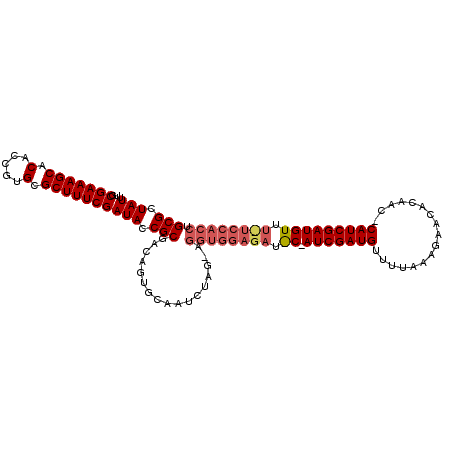
| Location | 13,695,014 – 13,695,124 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 66.87 |
| Mean single sequence MFE | -37.87 |
| Consensus MFE | -24.55 |
| Energy contribution | -28.00 |
| Covariance contribution | 3.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.84 |
| Structure conservation index | 0.65 |
| SVM decision value | 7.62 |
| SVM RNA-class probability | 1.000000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13695014 110 + 20766785 AGGUGGAAAAACAUCGAUGUGUUGUGUUCUUUAAAACAUCGAU-GUUUCUCCACCU-CUAGAUAGCAUUGUCGCGAUAUCGAAAGCACACGGUGUGCUUUCGUAAAUAGCGC (((((((.((((((((((((.(((.......))).))))))))-)))).)))))))-...((((....))))((.....(((((((((.....)))))))))......)).. ( -46.10) >DroSec_CAF1 6380 111 + 1 AGGUGGAGAAACAUCGAUGGGUUGUGUUCUUUAAAACAUCGAUGGCAUCUCCACCU-CUAGAUGGCACUGUCGCGGUAUCGAAAGCGCACGGUGUGCUUUCGCAAAUAGCGC (((((((((..((((((((..(((.......)))..))))))))...)))))))))-...(((......)))(((.((((((((((((.....)))))))))...))).))) ( -48.40) >DroSim_CAF1 6383 112 + 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNAUCUCCACCUCCUAGAUUGCACUGUCGCGGUAUCGAAAGCGCACGGUGUGCUUUCGCAAAUAGCGC ............................................................(((......)))(((.((((((((((((.....)))))))))...))).))) ( -19.10) >consensus AGGUGGA_AAACAUCGAUG_GUUGUGUUCUUUAAAACAUCGAU_G_AUCUCCACCU_CUAGAUAGCACUGUCGCGGUAUCGAAAGCGCACGGUGUGCUUUCGCAAAUAGCGC .((((((....(((((((((.(((.......))).))))))))).....)))))).....(((......)))(((.((((((((((((.....)))))))))...))).))) (-24.55 = -28.00 + 3.45)

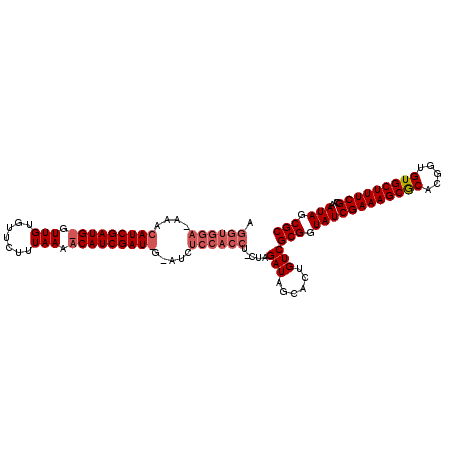
| Location | 13,695,014 – 13,695,124 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 66.87 |
| Mean single sequence MFE | -36.99 |
| Consensus MFE | -21.13 |
| Energy contribution | -24.33 |
| Covariance contribution | 3.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.12 |
| Structure conservation index | 0.57 |
| SVM decision value | 6.49 |
| SVM RNA-class probability | 0.999998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13695014 110 - 20766785 GCGCUAUUUACGAAAGCACACCGUGUGCUUUCGAUAUCGCGACAAUGCUAUCUAG-AGGUGGAGAAAC-AUCGAUGUUUUAAAGAACACAACACAUCGAUGUUUUUCCACCU (((.(((...(((((((((.....)))))))))))).)))...............-((((((((((((-((((((((...............)))))))))))))))))))) ( -47.86) >DroSec_CAF1 6380 111 - 1 GCGCUAUUUGCGAAAGCACACCGUGCGCUUUCGAUACCGCGACAGUGCCAUCUAG-AGGUGGAGAUGCCAUCGAUGUUUUAAAGAACACAACCCAUCGAUGUUUCUCCACCU (((((..((((((((((.(.....).))))))(....))))).))))).......-(((((((((...((((((((..((.........))..))))))))..))))))))) ( -41.50) >DroSim_CAF1 6383 112 - 1 GCGCUAUUUGCGAAAGCACACCGUGCGCUUUCGAUACCGCGACAGUGCAAUCUAGGAGGUGGAGAUNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN .((((...(((....)))..((.((((((.(((......))).)))))).....)).))))................................................... ( -21.60) >consensus GCGCUAUUUGCGAAAGCACACCGUGCGCUUUCGAUACCGCGACAGUGCAAUCUAG_AGGUGGAGAU_C_AUCGAUGUUUUAAAGAACACAAC_CAUCGAUGUUU_UCCACCU (((.(((...(((((((.(.....).)))))))))).))).................((((((((.((.(((((((.................))))))))).)))))))). (-21.13 = -24.33 + 3.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:20 2006