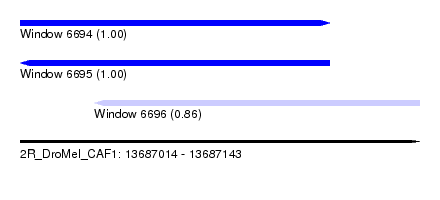
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,687,014 – 13,687,143 |
| Length | 129 |
| Max. P | 0.999865 |

| Location | 13,687,014 – 13,687,114 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 75.68 |
| Mean single sequence MFE | -24.02 |
| Consensus MFE | -12.22 |
| Energy contribution | -13.62 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.51 |
| SVM decision value | 3.37 |
| SVM RNA-class probability | 0.999089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13687014 100 + 20766785 UGUGCCAAACAUAGA------UAUGUACUAUGUACUUGCGUACAUAUGUAUGUAUUGUUUGUAUUUUGGCAAACCAAAACAAACAGAAAAUUUGCAUAUUAUGCGC (((((...(((((.(------(((((((((......)).)))))))).))))).((((((((..(((((....))))))))))))).......)))))........ ( -28.60) >DroSec_CAF1 2885 91 + 1 UGUGCCAAACAUAUA------UAUGUACUAUGUACUUGCGUACAUAUG---------UUUGUAUUUUGGCAAACCAAAACAAACAGAAAAUUUGCAUAUUAUGCGC .........((((.(------((((((.(((((((....)))))))((---------(((((..(((((....)))))))))))).......)))))))))))... ( -23.30) >DroSim_CAF1 2934 100 + 1 UGUGCCAAACAUACA------UAUGUACUAUGUACUUUCGAACAUAUGUAUGUCUUGUUUGUAUUUUGGCAAACCAAAACAAACAGAAAAUUUGCAUAUUAUGCGC (((((...(((((((------(((((.(...........).)))))))))))).((((((((..(((((....))))))))))))).......)))))........ ( -26.90) >DroYak_CAF1 2919 106 + 1 UGUGCCAAACACACAUAUACGUAUGUACCAUGUAAUUGCUUACGUAUGUAUGUAUUGUUUGUAUCUUGGCGAACCAAAACAAACAGAAAAUUUGCAUAUUAUGUGC (((((.........(((((((((((((.((......))..))))))))))))).((((((((...((((....)))).)))))))).......)))))........ ( -26.20) >DroAna_CAF1 2800 89 + 1 UUUGGCAAACAUA-----------------UGUACGCAGGUACAUAUAUACUUCUUAUAUGUAUGCAGGCAAACCAAAACAAAUACGAAAGCUUUAUUUCAGGUGG ((((.(....(((-----------------(((((....))))))))((((.........))))...).))))(((....((((((....)...)))))....))) ( -15.10) >consensus UGUGCCAAACAUACA______UAUGUACUAUGUACUUGCGUACAUAUGUAUGUAUUGUUUGUAUUUUGGCAAACCAAAACAAACAGAAAAUUUGCAUAUUAUGCGC (((((.......................(((((((....))))))).........(((((((..(((((....))))))))))))........)))))........ (-12.22 = -13.62 + 1.40)

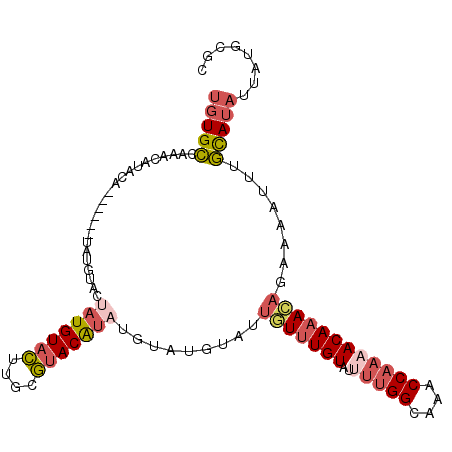
| Location | 13,687,014 – 13,687,114 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.68 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.80 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.64 |
| Structure conservation index | 0.62 |
| SVM decision value | 4.30 |
| SVM RNA-class probability | 0.999865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13687014 100 - 20766785 GCGCAUAAUAUGCAAAUUUUCUGUUUGUUUUGGUUUGCCAAAAUACAAACAAUACAUACAUAUGUACGCAAGUACAUAGUACAUA------UCUAUGUUUGGCACA ((((((((((((.........((((((((((((....)))))..))))))).....(((.(((((((....))))))))))))))------).)))))...))... ( -30.10) >DroSec_CAF1 2885 91 - 1 GCGCAUAAUAUGCAAAUUUUCUGUUUGUUUUGGUUUGCCAAAAUACAAA---------CAUAUGUACGCAAGUACAUAGUACAUA------UAUAUGUUUGGCACA ((((((((((((.........((((((((((((....)))))..)))))---------))(((((((....)))))))...))))------).)))))...))... ( -28.40) >DroSim_CAF1 2934 100 - 1 GCGCAUAAUAUGCAAAUUUUCUGUUUGUUUUGGUUUGCCAAAAUACAAACAAGACAUACAUAUGUUCGAAAGUACAUAGUACAUA------UGUAUGUUUGGCACA ((((((...))))........((((((((((((....)))))..)))))))((((((((((((((.(....)........)))))------))))))))).))... ( -31.10) >DroYak_CAF1 2919 106 - 1 GCACAUAAUAUGCAAAUUUUCUGUUUGUUUUGGUUCGCCAAGAUACAAACAAUACAUACAUACGUAAGCAAUUACAUGGUACAUACGUAUAUGUGUGUUUGGCACA ..........(((........((((((((((((....)))))..)))))))...((.((((((((((....))))...((((....))))..)))))).))))).. ( -26.60) >DroAna_CAF1 2800 89 - 1 CCACCUGAAAUAAAGCUUUCGUAUUUGUUUUGGUUUGCCUGCAUACAUAUAAGAAGUAUAUAUGUACCUGCGUACA-----------------UAUGUUUGCCAAA ....(..(((((((((....)).)))))))..).......(((.((((((.....))))((((((((....)))))-----------------))))).))).... ( -15.80) >consensus GCGCAUAAUAUGCAAAUUUUCUGUUUGUUUUGGUUUGCCAAAAUACAAACAAGACAUACAUAUGUACGCAAGUACAUAGUACAUA______UGUAUGUUUGGCACA ..(((((..............((((((((((((....)))))..))))))).........(((((((....)))))))...............)))))........ (-16.32 = -16.80 + 0.48)


| Location | 13,687,038 – 13,687,143 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.91 |
| Mean single sequence MFE | -22.55 |
| Consensus MFE | -7.91 |
| Energy contribution | -7.75 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13687038 105 - 20766785 C-CGCCACU---------CCA-AUGGAAACAAGUUAAUGUGCGCAUAAUAUGCAAAUUUUCUGUUUGUUUUGGUUUGCCAAAAUACAAACAAUACAUACAUAUGUACGCAAGUACA .-.(((((.---------...-.((....)).......))).))...(((((.........((((((((((((....)))))..))))))).......)))))((((....)))). ( -26.19) >DroPse_CAF1 2596 101 - 1 C-CGUCCGUUCGCAAUUUGCA-AGCAAAACAAA-AAAUG-GCGCAAGACA-ACAAAUGUUGUGUUUGUUUC-GUUUGUCAAGUUU---AU----CAUAUGUAUGUA--CAUGUACA .-...........((((((((-(((.(((((((-.....-.(....)(((-((....))))).))))))).-))))).)))))).---..----.((((((....)--)))))... ( -22.50) >DroSec_CAF1 2909 96 - 1 C-CGCCACU---------CCA-AUGAAAACAAGUUAAUGUGCGCAUAAUAUGCAAAUUUUCUGUUUGUUUUGGUUUGCCAAAAUACAAA---------CAUAUGUACGCAAGUACA .-.(((((.---------..(-((........)))...))).)).................((((((((((((....)))))..)))))---------))..(((((....))))) ( -22.50) >DroSim_CAF1 2958 106 - 1 CCCGCCACU---------CCA-AUGGAAACAAGUUAAUGUGCGCAUAAUAUGCAAAUUUUCUGUUUGUUUUGGUUUGCCAAAAUACAAACAAGACAUACAUAUGUUCGAAAGUACA .........---------...-.((....))......(((((((((...))))....((((((((((((((((....)))))..))))))).(((((....))))).))))))))) ( -23.70) >DroYak_CAF1 2949 105 - 1 C-CGCCACU---------UCA-AUAGAAUCAAGGAAAUAUGCACAUAAUAUGCAAAUUUUCUGUUUGUUUUGGUUCGCCAAGAUACAAACAAUACAUACAUACGUAAGCAAUUACA .-.......---------...-.........(((((((.((((.......)))).)))))))(((((((((((....)))))..)))))).............((((....)))). ( -21.50) >DroPer_CAF1 2584 102 - 1 C-CGUCCGUUCGCAAUUUGCAAAGCAAAACAAA-AAAUG-GCGCAAGACA-ACAAAUGUUGUGUUUGUUUC-GUUUGUCAAGUUU---AU----CAUAUGUAUGUA--CAUGUACA .-...........((((((..((((.(((((((-.....-.(....)(((-((....))))).))))))).-))))..)))))).---..----.((((((....)--)))))... ( -18.90) >consensus C_CGCCACU_________CCA_AUGAAAACAAGUAAAUGUGCGCAUAAUAUGCAAAUUUUCUGUUUGUUUUGGUUUGCCAAAAUACAAAC____CAUACAUAUGUACGCAAGUACA ..........................................(((.(((..((((((.....))))))....)))))).........................((((....)))). ( -7.91 = -7.75 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:16 2006