
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,682,740 – 13,682,851 |
| Length | 111 |
| Max. P | 0.638534 |

| Location | 13,682,740 – 13,682,851 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.32 |
| Mean single sequence MFE | -32.16 |
| Consensus MFE | -22.67 |
| Energy contribution | -22.53 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13682740 111 + 20766785 CGAAAUCAGCUGAAGCUUAAGCAGCAGCUAAUUCGCAAAUACAAGGACACAGAGUUCUAGGCGGC---AGCA-AAAUAAAA--AAAAGGGCAAGAUGGUGCUGCGACUGCUAAUGCG .(((...(((((..((....))..)))))..)))(((......(((((.....))))).((((((---((((-..((....--...........))..))))))...))))..))). ( -28.46) >DroVir_CAF1 224 111 + 1 CGCAGUCAGCUGAAGCUGAAGCAGCAGCUCAUACGCAAAUACAAGGAUACGGAGUUCUAGGAUAC---ACAC-UCACACAC--ACACUGACAGGAUGGUGCUGCGACUGCUGAUGCG ((((.((((((((.((((......)))))))..((((....(.(((((.....))))).).....---....-...(((.(--(..(.....)..))))).))))...))))))))) ( -28.50) >DroSec_CAF1 565 108 + 1 CGAAAUCAGCUGAAGCUUAAGCAGCAGCUAAUUCGCAAGUACAAGGACACAGAGUUCUAGGCGGC---AGCG-AAAUAAGA-----AAGGCAAGAUGGUGCUGCGACUGCUAAUGCG .(((...(((((..((....))..)))))..)))(((.((...(((((.....)))))..))(((---((((-........-----..((((......)))).)).)))))..))). ( -29.30) >DroEre_CAF1 557 115 + 1 CGAAAUCAGCUUAAGCUCAAGCAGCAGCUGAUCCGCAAGUACAAGGACACAGAGUUCUAGGCAGUGGUAGUACAAAAAAAG--AGUCGGGCAAGAUGGUGCUGCGGCUGCUGAUGCG ....(((((((...((....))...))))))).((((......(((((.....))))).((((((.(((((((........--.(((......))).))))))).))))))..)))) ( -39.20) >DroYak_CAF1 873 114 + 1 CGAAAUCAGCUUAAGCUCAAGCAGCAGCUGAUUCGCAAGUACAAGGACACAGAGUUCUAGGCGGUGGUAGUA-AAACAAGG--AGCCGGGCAAGAUGGUGCUGCGACUGCUGAUGCG .(((.((((((...((....))...)))))))))(((......(((((.....))))).((((((.((((((-...((...--.((...))....)).)))))).))))))..))). ( -35.90) >DroAna_CAF1 568 108 + 1 CGCAACCAGCUGAAGCUCAAGCAGCAGCUCAUUCGCAAGUACAAGGACAUAGAGUUCUAG--------AGCA-GAUUCGGGAGAGCAAAACAAGAUGGUGCUGCGACUGCUGAUGCG ((((..((((((.....)).((((((((((...(....)....(((((.....))))).)--------))).-(.(((....))))............))))))....)))).)))) ( -31.60) >consensus CGAAAUCAGCUGAAGCUCAAGCAGCAGCUAAUUCGCAAGUACAAGGACACAGAGUUCUAGGCGGC___AGCA_AAAUAAAG__AGAAGGGCAAGAUGGUGCUGCGACUGCUGAUGCG ........((...(((....((((((((......)).......(((((.....)))))........................................))))))....)))...)). (-22.67 = -22.53 + -0.14)

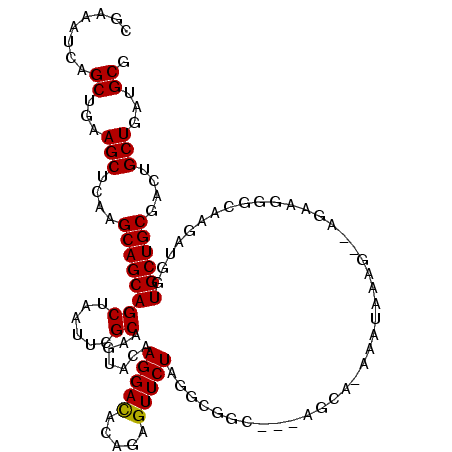
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:12 2006