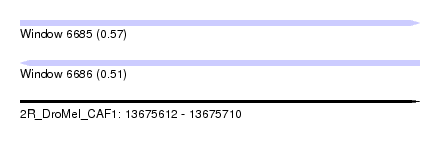
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,675,612 – 13,675,710 |
| Length | 98 |
| Max. P | 0.572076 |

| Location | 13,675,612 – 13,675,710 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.16 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -14.38 |
| Energy contribution | -16.13 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
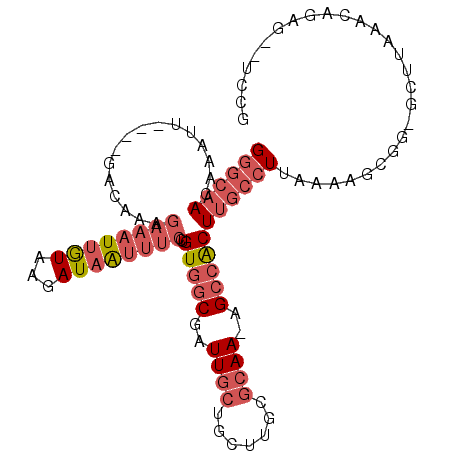
>2R_DroMel_CAF1 13675612 98 + 20766785 GGGCAACAAAUU----GACAAAGAAAUUGUAAGAUAAUUUCCUGUGGCGAUUGCUGCUUGCGCAA-AGCCACUUGCCUUGAAAGCGG-GCUUAAACAGAG--UCCG ((((((......----......((((((((...))))))))..(((((..((((.......))))-.)))))))))))......(((-((((.....)))--)))) ( -30.80) >DroSec_CAF1 5545 98 + 1 GGGCAACAAAUU----GACAAAGAAAUUGUAAGAUAAUUUCCUGUGGCUAUUGCUGGUUGCGCAA-AGCCACUUGCCUUAAAAGCGG-GCUUAAACGGAG--UCCU ((((((......----......((((((((...))))))))..((((((.((((.......))))-)))))))))))).......((-((((.....)))--))). ( -30.80) >DroSim_CAF1 5728 98 + 1 GGGCAACAAAUU----GACAAAGAAAUUGUAAGAUAAUUUCCUGUGGCUAUUGCUGGUUGUGCAA-AGCCACUUGCCUUAAAAGCGG-GCUUAAACGGAG--UCCG ((((((......----......((((((((...))))))))..((((((.((((.......))))-))))))))))))......(((-((((.....)))--)))) ( -32.50) >DroEre_CAF1 5628 99 + 1 GGGCAACAAAUU----GACAAAGAAAUUGUAAGAUAAUUUCCUGUGGCGAUUGCUGGUUGCGCAAAAGCCACUUGCCUCAAAAGCGG-GCUCAAACAGAG--UCCG ((((((......----......((((((((...))))))))..(((((..((((.......))))..)))))))))))......(((-((((.....)))--)))) ( -33.70) >DroYak_CAF1 5669 99 + 1 GGGCAACAAAUU----GACAAAGAAAUUGUAAGAUAGUUUCCUGUGGCGAUUGCUGCUUGCGCAAAAGCCACUUGCCUCAAAAGCGG-GCUUAAACAGAG--UCCG ((((((......----......((((((((...))))))))..(((((..((((.......))))..)))))))))))......(((-((((.....)))--)))) ( -31.60) >DroAna_CAF1 5548 97 + 1 GACAAAGAAUUUGUGUGGCACAGAAUUUAUUAAAUUA------GUAACAAUUGCAGCAACCUAAAAUGCCGCUGGUCU--AAGGCAGAGAUUACA-AGUGUUCCCA .((((.....))))(..((((..(((((......(((------(...((...((.(((........))).))))..))--)).....)))))...-.))))..).. ( -13.40) >consensus GGGCAACAAAUU____GACAAAGAAAUUGUAAGAUAAUUUCCUGUGGCGAUUGCUGCUUGCGCAA_AGCCACUUGCCUUAAAAGCGG_GCUUAAACAGAG__UCCG ((((((................((((((((...))))))))..(((((..((((.......))))..)))))))))))............................ (-14.38 = -16.13 + 1.75)

| Location | 13,675,612 – 13,675,710 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.16 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -14.62 |
| Energy contribution | -16.65 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
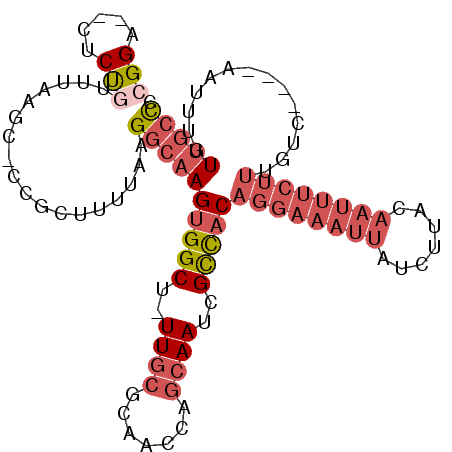
>2R_DroMel_CAF1 13675612 98 - 20766785 CGGA--CUCUGUUUAAGC-CCGCUUUCAAGGCAAGUGGCU-UUGCGCAAGCAGCAAUCGCCACAGGAAAUUAUCUUACAAUUUCUUUGUC----AAUUUGUUGCCC (((.--((.......)).-))).......((((((((((.-((((....)))).....)))))((((((((.......))))))))....----......))))). ( -25.80) >DroSec_CAF1 5545 98 - 1 AGGA--CUCCGUUUAAGC-CCGCUUUUAAGGCAAGUGGCU-UUGCGCAACCAGCAAUAGCCACAGGAAAUUAUCUUACAAUUUCUUUGUC----AAUUUGUUGCCC .((.--((.......)).-))........(((((((((((-((((.......)))).))))))((((((((.......))))))))....----......))))). ( -25.80) >DroSim_CAF1 5728 98 - 1 CGGA--CUCCGUUUAAGC-CCGCUUUUAAGGCAAGUGGCU-UUGCACAACCAGCAAUAGCCACAGGAAAUUAUCUUACAAUUUCUUUGUC----AAUUUGUUGCCC (((.--((.......)).-))).......(((((((((((-((((.......)))).))))))((((((((.......))))))))....----......))))). ( -26.00) >DroEre_CAF1 5628 99 - 1 CGGA--CUCUGUUUGAGC-CCGCUUUUGAGGCAAGUGGCUUUUGCGCAACCAGCAAUCGCCACAGGAAAUUAUCUUACAAUUUCUUUGUC----AAUUUGUUGCCC (((.--(((.....))).-))).......((((((((((..((((.......))))..)))))((((((((.......))))))))....----......))))). ( -31.00) >DroYak_CAF1 5669 99 - 1 CGGA--CUCUGUUUAAGC-CCGCUUUUGAGGCAAGUGGCUUUUGCGCAAGCAGCAAUCGCCACAGGAAACUAUCUUACAAUUUCUUUGUC----AAUUUGUUGCCC (((.--((.......)).-))).......((((((((((..((((.......))))..)))))((....)).....((((.....)))).----......))))). ( -25.80) >DroAna_CAF1 5548 97 - 1 UGGGAACACU-UGUAAUCUCUGCCUU--AGACCAGCGGCAUUUUAGGUUGCUGCAAUUGUUAC------UAAUUUAAUAAAUUCUGUGCCACACAAAUUCUUUGUC .((((((...-.))..)))).((..(--(((...((((((........))))))..((((((.------.....))))))..)))).))................. ( -15.70) >consensus CGGA__CUCUGUUUAAGC_CCGCUUUUAAGGCAAGUGGCU_UUGCGCAACCAGCAAUCGCCACAGGAAAUUAUCUUACAAUUUCUUUGUC____AAUUUGUUGCCC (((.....)))..................((((((((((..((((.......))))..)))))((((((((.......))))))))..............))))). (-14.62 = -16.65 + 2.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:06 2006