
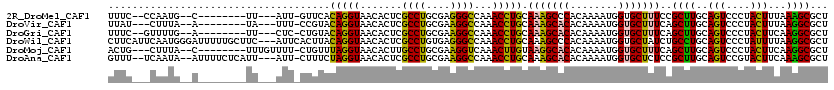
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,670,274 – 13,670,378 |
| Length | 104 |
| Max. P | 0.656331 |

| Location | 13,670,274 – 13,670,378 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.22 |
| Mean single sequence MFE | -25.75 |
| Consensus MFE | -18.64 |
| Energy contribution | -17.95 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
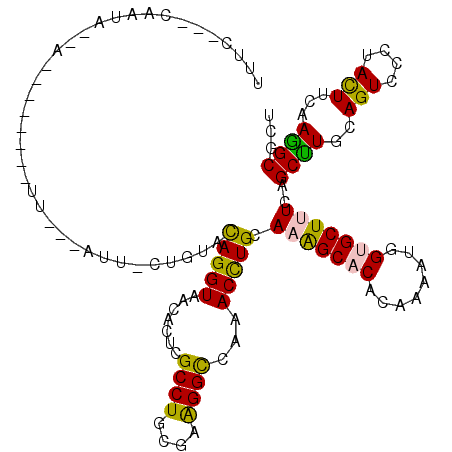
>2R_DroMel_CAF1 13670274 104 - 20766785 UUUC--CCAAUG--C--------UU---AUU-GUUCACAGGUAACACUCGCCUGCGAGGGCCAAACCUGCAAAGCCCACAAAAUGGUGCUUUCCGCUUGCAGUCCCUACUUUAAAGCGCU ....--.....(--(--------((---...-..((.(((((.......))))).))(((......((((((((((((.....))).)))......)))))).))).......))))... ( -25.10) >DroVir_CAF1 262 103 - 1 UUAU---CUUUA--A--------UA---UUU-CCGUACAGGUAACACUCGCCUGCGAAGGCCAAACCUGCAAAGCACACAAAAUGGUGCUUUCAGCUUGCAGUCCCUACUUUAAGGCGCU ((((---((.((--.--------..---...-...)).))))))....(((((..((((.......(((((((((((........)))))......))))))......)))).))))).. ( -23.22) >DroGri_CAF1 270 104 - 1 UUUC--GUUUUG--A--------UU---CUC-CUGUACAGGUAACACUCGCCUGCGAAGGCCAAACCUGCAAAGCACACAAAAUGGUGCUUUCAGCUUGCAGUCCCUACUUCAAGGCGCU ...(--((((((--(--------..---...-((((((((((.......((((....))))...))))).(((((((........))))))).....)))))........)))))))).. ( -28.22) >DroWil_CAF1 339 117 - 1 CUUCAUUCAAUGGGAUUUUUGCUUC---AUUCACUUACAGGUAACACUCGCCUGUGAGGGCCAAACCUGCAAAGCCCACAAAAUGGUGCUAUCUGCCUGCAGUCCCUAUUUUAAGGCGCU ...((((...((((...(((((...---.....(((((((((.......)))))))))((.....)).))))).))))...))))(((((..(((....)))............))))). ( -30.94) >DroMoj_CAF1 262 106 - 1 ACUG---CUUUA--C--------UUUGUUUU-CUGUUUAGGUAACACUUGCCUGCGAAGGUCAAACUUGUAAGGCACACAAAAUGGUGCUUUCAGCUUGCAGUCCCUACUUCAAGGCGCU ..((---(((((--(--------...(((((-((...((((((.....))))))...)))..))))..))))))))........(((((......((((.(((....))).))))))))) ( -25.30) >DroAna_CAF1 261 112 - 1 GUUU--UCAAUA--AUUUUCUCAUU---AUU-CUUUCUAGGUAACACUCGCCUGCGAAGGCCAAACCUGCAAAGCACACAAAAUGGUGCUCUCCGCUUGCAGUCCGUACUUCAAAGCGCU ((((--(.....--...........---...-((((((((((.......))))).)))))......(((((((((((........)))))......))))))..........)))))... ( -21.70) >consensus UUUC___CAAUA__A________UU___AUU_CUGUACAGGUAACACUCGCCUGCGAAGGCCAAACCUGCAAAGCACACAAAAUGGUGCUUUCAGCUUGCAGUCCCUACUUCAAGGCGCU .....................................(((((.......((((....))))...))))).(((((((........)))))))..((((..(((....)))...))))... (-18.64 = -17.95 + -0.69)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:03 2006