| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,659,797 – 13,659,901 |
| Length | 104 |
| Max. P | 0.749409 |

| Location | 13,659,797 – 13,659,901 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 75.86 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -17.24 |
| Energy contribution | -18.13 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
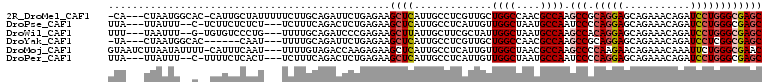
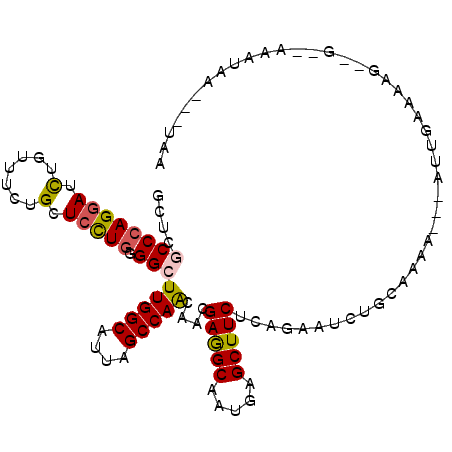
>2R_DroMel_CAF1 13659797 104 + 20766785 -CA---CUAAUGGCAC-CAUUGCUAUUUUUCUUGCAGAUUCUGAGAAGCUCAUUGCCUCGUUGCUGGCCAACGCCAAGCCGCAGGAGCAGAAACAGAUCCUGGGCGAGC -..---..(((((((.-...))))))).(((((.........)))))((((.............((((....)))).(((.(((((...........)))))))))))) ( -29.00) >DroPse_CAF1 17088 100 + 1 UUA---UUAUUU--C-UCUUCUCUCU---UCUUUCAGACUCUGAGAAGCUCAUUGCCUCAUUGUUGGCUAAUGCCAAUCCCCAGGAGCAGAAACAGAUCCUGGGCGAGC ...---....((--(-((...(((..---......)))....)))))((((...........((((((....)))))).(((((((...........))))))).)))) ( -29.10) >DroWil_CAF1 18674 100 + 1 UUU---UAAUUU--G-UGUGUCCCUG---UUUUGCAGAUCCCGAGAAGCUUAUUGCUUCGCUAUUGGCUAAUGCCAAGCCACAGGAGCAGAAACAGAUCCUGGGCGAGC ...---......--.-.......(((---.....))).......(((((.....)))))(((.(((((....)))))(((.(((((...........)))))))).))) ( -30.30) >DroYak_CAF1 16794 96 + 1 -UA---CUAAUGGCAC------CAAU---UUUUGCAGAUUCUGAGAAGCUCAUUGCCUCGUUGCUGGCCAACGCCAAGCCGCAGGAGCAGAAACAGAUCCUCGGCGAGC -..---....((((..------....---....((((....((((...))))))))...((((.....)))))))).((((.((((...........)))))))).... ( -24.20) >DroMoj_CAF1 17966 105 + 1 GUAAUCUUAAUAUUUU-CAUUUCAAU---UUUUGUAGACCAAGAGAAGCUCAUUGCCUCAUUGUUGGCUAACGCCAAGCCCCAAGAACAGAAACAAAUUCUGGGCGAAC ................-........(---(((((.....)))))).......(((((......(((((....)))))..........(((((.....)))))))))).. ( -20.10) >DroPer_CAF1 16349 100 + 1 UUA---UUAUUU--C-UUUUCUCACU---UCUUUCAGACUCUGAGAAGCUCAUUGCCUCAUUGUUGGCUAAUGCCAAUCCCCAGGAGCAGAAACAGAUCCUGGGCGAGC ...---......--.-..((((((..---((.....))...))))))((((...........((((((....)))))).(((((((...........))))))).)))) ( -29.70) >consensus UUA___UUAAUU__A__CUUUUCAAU___UCUUGCAGACUCUGAGAAGCUCAUUGCCUCAUUGUUGGCUAACGCCAAGCCCCAGGAGCAGAAACAGAUCCUGGGCGAGC ...............................................((((.............((((....)))).(((.(((((...........)))))))))))) (-17.24 = -18.13 + 0.89)
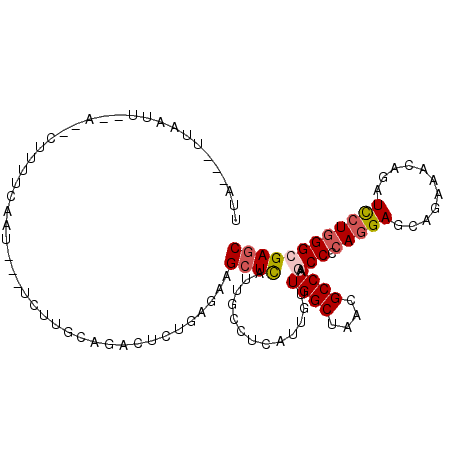
| Location | 13,659,797 – 13,659,901 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 75.86 |
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -20.00 |
| Energy contribution | -20.03 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13659797 104 - 20766785 GCUCGCCCAGGAUCUGUUUCUGCUCCUGCGGCUUGGCGUUGGCCAGCAACGAGGCAAUGAGCUUCUCAGAAUCUGCAAGAAAAAUAGCAAUG-GUGCCAUUAG---UG- ((..((((((((.(.......).))))).))).((((....)))))).....((((.((((...)))).....(((..........)))...-.)))).....---..- ( -28.80) >DroPse_CAF1 17088 100 - 1 GCUCGCCCAGGAUCUGUUUCUGCUCCUGGGGAUUGGCAUUAGCCAACAAUGAGGCAAUGAGCUUCUCAGAGUCUGAAAGA---AGAGAGAAGA-G--AAAUAA---UAA ((((((((((((.(.......).)))))))..(((((....)))))...........)))))(((((....(((......---))).....))-)--))....---... ( -30.20) >DroWil_CAF1 18674 100 - 1 GCUCGCCCAGGAUCUGUUUCUGCUCCUGUGGCUUGGCAUUAGCCAAUAGCGAAGCAAUAAGCUUCUCGGGAUCUGCAAAA---CAGGGACACA-C--AAAUUA---AAA .........(..(((((((.((((((((..(((((((....))))..)))(((((.....))))).)))))...))))))---))))..)...-.--......---... ( -31.20) >DroYak_CAF1 16794 96 - 1 GCUCGCCGAGGAUCUGUUUCUGCUCCUGCGGCUUGGCGUUGGCCAGCAACGAGGCAAUGAGCUUCUCAGAAUCUGCAAAA---AUUG------GUGCCAUUAG---UA- ....((((((((.(.......).)))).)))).((((((((.....))))(((((.....)))))...............---....------..))))....---..- ( -27.10) >DroMoj_CAF1 17966 105 - 1 GUUCGCCCAGAAUUUGUUUCUGUUCUUGGGGCUUGGCGUUAGCCAACAAUGAGGCAAUGAGCUUCUCUUGGUCUACAAAA---AUUGAAAUG-AAAAUAUUAAGAUUAC .((((((((((((........)))))..))))(((((....)))))(((((((((.....)))))..(((.....)))..---))))....)-)).............. ( -24.50) >DroPer_CAF1 16349 100 - 1 GCUCGCCCAGGAUCUGUUUCUGCUCCUGGGGAUUGGCAUUAGCCAACAAUGAGGCAAUGAGCUUCUCAGAGUCUGAAAGA---AGUGAGAAAA-G--AAAUAA---UAA ((((((((((((.(.......).)))))))..(((((....)))))...........)))))((((((...((.....))---..))))))..-.--......---... ( -31.30) >consensus GCUCGCCCAGGAUCUGUUUCUGCUCCUGGGGCUUGGCAUUAGCCAACAACGAGGCAAUGAGCUUCUCAGAAUCUGCAAAA___AUUGAAAAG__G__AAAUAA___UAA ....((((((((.(.......).))))).)))(((((....)))))....(((((.....)))))............................................ (-20.00 = -20.03 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:01 2006