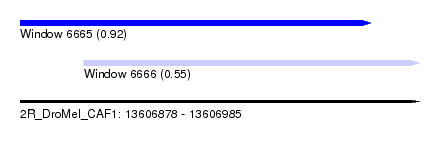
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,606,878 – 13,606,985 |
| Length | 107 |
| Max. P | 0.919076 |

| Location | 13,606,878 – 13,606,972 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.87 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -14.97 |
| Energy contribution | -14.80 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
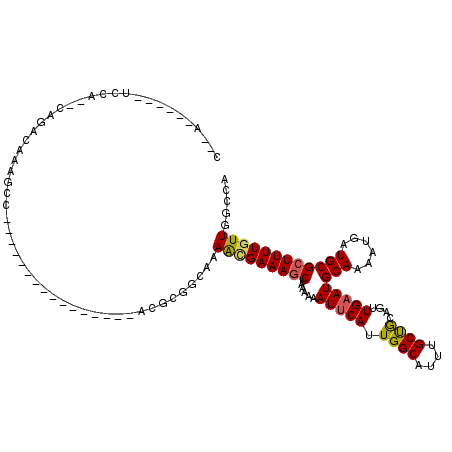
>2R_DroMel_CAF1 13606878 94 + 20766785 C--A------UCCA--CAGACAAAGCC----------------GCGCGCCAAAGCGAAAGCUAAAAAAUUCAUUGGCAUUUGCUGCAGUUGAAUGCAAAAUGAUGCGCUUUUGUUGGCCA .--.------....--..(((((((..----------------(((((.((.((((((.(((((........))))).))))))(((......)))....)).))))))))))))..... ( -26.30) >DroVir_CAF1 158516 112 + 1 GCGA------CGGG--CAGACAAAGCGCGCACGCGGAUGGCAAACGCCGCGAAAUGAAAGCUAAAAAAUGCAUUGGCAUUUGCUGCAGUUGAAUGCAAAAUGAUGCGCUUUUGUUGCCGA ....------..((--(((..(((((((((.(((((.((.....)))))))..............((((((....))))))))((((......)))).......)))))))..))))).. ( -39.20) >DroPse_CAF1 146230 91 + 1 C--A------AUCC--CGAACAAAGCC----------------GCGC---AAAAAGAAAGCUAAAAAAUUCAUUGGCAUUUGCCGCAGUUGAAUGCAAAAUGAUGCGCUUUUGUUGGCCA .--.------...(--(.(((((((..----------------((((---(...((....)).......((((((((....)))(((......)))..)))))))))))))))))))... ( -22.70) >DroGri_CAF1 136775 101 + 1 CAAA------UAGG--CGGACAAAGCGC-----------GCAAACGCCGCAAAAUGAAAGCUAAAAAAUUCAUUGGCAUUUGCUGCAGUUGAAUGCAAAAUGAUGCGCUUUUGUUGGCCA ....------..((--(.((((((((((-----------(((((.((((....(((((..........))))))))).)))))((((......)))).......)))).)))))).))). ( -36.80) >DroWil_CAF1 170545 92 + 1 G--A------UGUC--AGUACAAAGCC----------------C--AGGCAAAACGAAAACUAAAAAAUUCAUUGGCAUUUGCUACAGUUGAAUGCAAAAUGAUGCGCUUUUGUUGGCCA .--.------....--...........----------------.--.(((..(((((((.(......(((((.((((....))))....)))))(((......)))).))))))).))). ( -17.30) >DroAna_CAF1 146301 102 + 1 C--AAUCUCCUCCAGGCAGACAAAGCU----------------UCGCGGCAAAGCGAAAGCUAAAAAAUUCAUUGGCAUUUGCUGCAGUUGAAUGCAAAAUGAUGCGCUUUUGUUGGCCA .--...........(((.(((((((..----------------.((((.((.((((((.(((((........))))).))))))(((......)))....)).)))).))))))).))). ( -29.80) >consensus C__A______UCCA__CAGACAAAGCC________________ACGCGGCAAAACGAAAGCUAAAAAAUUCAUUGGCAUUUGCUGCAGUUGAAUGCAAAAUGAUGCGCUUUUGUUGGCCA ....................................................(((((((((......(((((.((((....))))....)))))(((......))))))))))))..... (-14.97 = -14.80 + -0.17)

| Location | 13,606,895 – 13,606,985 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 81.84 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -14.47 |
| Energy contribution | -14.38 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13606895 90 + 20766785 ---GCGCGCCAAAGCGAAAGCUAAAAAAUUCAUUGGCAUUUGCUGCAGUUGAAUGCAAAAUGAUGCGCUUUUGUUGGCCAACUGGAAAAAUUG-------- ---..(.((((..((((((((......(((((...(((.....)))...)))))(((......))))))))))))))))..............-------- ( -24.30) >DroVir_CAF1 158548 94 + 1 CAAACGCCGCGAAAUGAAAGCUAAAAAAUGCAUUGGCAUUUGCUGCAGUUGAAUGCAAAAUGAUGCGCUUUUGUUGCCGAAGUGCAAACAAAAA------- ....(((..((.((..(((((....((((((....))))))..((((......)))).........)))))..))..))..)))..........------- ( -21.10) >DroPse_CAF1 146247 90 + 1 ---GCGC---AAAAAGAAAGCUAAAAAAUUCAUUGGCAUUUGCCGCAGUUGAAUGCAAAAUGAUGCGCUUUUGUUGGCCAGCUGGAAAAAGGGACU----- ---((.(---((...((((((......(((((.((((....))))....)))))(((......))))))))).)))))..................----- ( -19.70) >DroGri_CAF1 136796 98 + 1 CAAACGCCGCAAAAUGAAAGCUAAAAAAUUCAUUGGCAUUUGCUGCAGUUGAAUGCAAAAUGAUGCGCUUUUGUUGGCCAGCUGCAAACAAAAA---AAUG ((((.((((....(((((..........))))))))).)))).((((((((...(((......)))(((......)))))))))))........---.... ( -23.20) >DroWil_CAF1 170562 86 + 1 ---C--AGGCAAAACGAAAACUAAAAAAUUCAUUGGCAUUUGCUACAGUUGAAUGCAAAAUGAUGCGCUUUUGUUGGCCAGCUGGAA--ACUG-------- ---.--.(((..(((((((.(......(((((.((((....))))....)))))(((......)))).))))))).)))....(...--.)..-------- ( -18.30) >DroMoj_CAF1 191408 101 + 1 CAAACGCCGCAAAAUGAAAGCUAAAAAAUUCAUUGGCAUUUGCUGCAGUUGAAUGCAAAAUGAUGCGCUUUUGUUGCCCAGCUGCAAAAAAAAAGUAAAAG ((((.((((....(((((..........))))))))).)))).((((((((...(((......)))((.......)).))))))))............... ( -21.80) >consensus ___ACGCCGCAAAAUGAAAGCUAAAAAAUUCAUUGGCAUUUGCUGCAGUUGAAUGCAAAAUGAUGCGCUUUUGUUGGCCAGCUGCAAAAAAAAA_______ ............(((((((((......(((((.((((....))))....)))))(((......)))))))))))).......................... (-14.47 = -14.38 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:47 2006