| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,600,121 – 13,600,212 |
| Length | 91 |
| Max. P | 0.695715 |

| Location | 13,600,121 – 13,600,212 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -30.84 |
| Consensus MFE | -20.74 |
| Energy contribution | -21.48 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13600121 91 + 20766785 ACAUGGGGCUGUGUAAACGGCUGAGCGAGUGUGCGUGUGCUGAUUGCAUUGUUGCACUU---------GUUGCAUUCCCACCACGCACACACUCUCAAGC ......((((((....))))))(((.(.(((((((((((..((.((((...........---------..)))).)).)).))))))))).).))).... ( -33.52) >DroSec_CAF1 147800 91 + 1 ACAUGGCGUUGUGUAAACGGCUGAGCGAGUGUGCGUGUGCUGAUUGCAUUGUUGCACUU---------GUUGCAUUCCCUCCACGCACACACUCUCAAGC ....(.((((.....)))).)((((.(.((((((((((((.....)))....((((...---------..)))).......))))))))).).))))... ( -30.90) >DroSim_CAF1 146171 91 + 1 ACAUGGGGUUGUGUAAACGGCUGAGCGAGUGUGCGUGUGCUGAUUGCAUUGUUGCACUU---------GUUGCAUUCCCUCCACGCACACACUCUCAAGC ......((((((....))))))(((.(.((((((((((((.....)))....((((...---------..)))).......))))))))).).))).... ( -30.60) >DroEre_CAF1 143183 91 + 1 ACAUGGGGUUGUGUAAACGGCUGAGUGAGUGUGCGUGUGCUGAUUGCAUUGUUGCACUU---------GUUGCAUUCCCUCCACACACACGCUCGCAAAU ......((((((....))))))..(((((((((.(((((..((.((((...........---------..)))).))....))))).))))))))).... ( -32.72) >DroYak_CAF1 147538 91 + 1 ACAUGGGGUUGUGUAAACGGCUGAGCGUGUGUGCGUGUGCUGAUUGCAUUGUUGCACUU---------GUUGCAUUCCCUCCACGCACACGCUCGCAAAC ......((((((....))))))(((((((((((.(.((((.((.((((....)))).))---------...))))......).)))))))))))...... ( -32.20) >DroAna_CAF1 140220 91 + 1 AUAUACUG-UGUGUUAGUG-CU-AGUAUAUGAGUGUGCAUUGAUUGUAUUGUUGCACUUGUUGCACUUGUUGCAUUCCUCGCACGCACACU--CCC---- .......(-(((((..(((-(.-((...(((...(((((..((.((((....)))).))..)))))......)))..)).)))))))))).--...---- ( -25.10) >consensus ACAUGGGGUUGUGUAAACGGCUGAGCGAGUGUGCGUGUGCUGAUUGCAUUGUUGCACUU_________GUUGCAUUCCCUCCACGCACACACUCUCAAGC ......((((((....))))))(((...(((((((((....((.((((......................)))).))....))))))))).)))...... (-20.74 = -21.48 + 0.74)

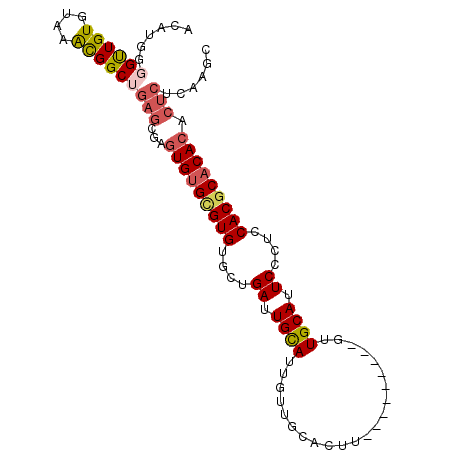
| Location | 13,600,121 – 13,600,212 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -20.08 |
| Energy contribution | -21.25 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13600121 91 - 20766785 GCUUGAGAGUGUGUGCGUGGUGGGAAUGCAAC---------AAGUGCAACAAUGCAAUCAGCACACGCACACUCGCUCAGCCGUUUACACAGCCCCAUGU (((.(((.(.(((((((((((..((.((((..---------...........)))).)).)).))))))))).).))))))................... ( -30.82) >DroSec_CAF1 147800 91 - 1 GCUUGAGAGUGUGUGCGUGGAGGGAAUGCAAC---------AAGUGCAACAAUGCAAUCAGCACACGCACACUCGCUCAGCCGUUUACACAACGCCAUGU (((.(((.(.(((((((((.......((((..---------...))))....(((.....)))))))))))).).))))))((((.....))))...... ( -30.70) >DroSim_CAF1 146171 91 - 1 GCUUGAGAGUGUGUGCGUGGAGGGAAUGCAAC---------AAGUGCAACAAUGCAAUCAGCACACGCACACUCGCUCAGCCGUUUACACAACCCCAUGU (((.(((.(.(((((((((.......((((..---------...))))....(((.....)))))))))))).).))))))................... ( -30.00) >DroEre_CAF1 143183 91 - 1 AUUUGCGAGCGUGUGUGUGGAGGGAAUGCAAC---------AAGUGCAACAAUGCAAUCAGCACACGCACACUCACUCAGCCGUUUACACAACCCCAUGU ......(((.(((((((((....((.((((..---------...........)))).))..))))))))).))).......................... ( -23.62) >DroYak_CAF1 147538 91 - 1 GUUUGCGAGCGUGUGCGUGGAGGGAAUGCAAC---------AAGUGCAACAAUGCAAUCAGCACACGCACACACGCUCAGCCGUUUACACAACCCCAUGU ....(((((((((((((((.......((((..---------...))))....(((.....))))))))))))..)))).))................... ( -28.40) >DroAna_CAF1 140220 91 - 1 ----GGG--AGUGUGCGUGCGAGGAAUGCAACAAGUGCAACAAGUGCAACAAUACAAUCAAUGCACACUCAUAUACU-AG-CACUAACACA-CAGUAUAU ----..(--(((((((((.....((.((.......((((.....))))......)).)).))))))))))(((((((-..-..........-.))))))) ( -23.14) >consensus GCUUGAGAGUGUGUGCGUGGAGGGAAUGCAAC_________AAGUGCAACAAUGCAAUCAGCACACGCACACUCGCUCAGCCGUUUACACAACCCCAUGU ..(((((.(.(((((((((....((.((((......................)))).))....))))))))).).))))).................... (-20.08 = -21.25 + 1.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:44 2006