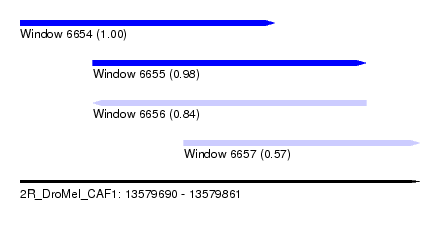
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,579,690 – 13,579,861 |
| Length | 171 |
| Max. P | 0.996693 |

| Location | 13,579,690 – 13,579,799 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 87.92 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -25.76 |
| Energy contribution | -25.82 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13579690 109 + 20766785 AUGGCAAGUGCAGCUUCCGCUGGAACUAAAACGGAAACAAAAAUGAAAAUGAAAAUGCAACUGAAACUGAAACUGGAACUCAAACGGCAGCCGAAGCUGCGACUGUAAC ...(((..(((((((((.((((..........(....)..................................(((.........))))))).)))))))))..)))... ( -27.90) >DroSec_CAF1 132746 109 + 1 AUGGCAAGUGCAGCUUCCGCUGGAACUAAAACGGAAACAAAAAUGAAAAGGAAAAUGCAACUGAAACUGUAACUGGAACUCAAACGGCAGCCGAAGCUGCGACUGUAAC ...(((..(((((((((.((((..........(....).................((((........))))................)))).)))))))))..)))... ( -28.60) >DroSim_CAF1 131124 109 + 1 AUGGCAAGUGCAGCUUCCGCUGGAACUAAAACGGAAACAAAAAUGAAAAUGAAAAUGCAACUGAAACUGUAACUGGAACUCAAACGGCAGCCGAAGCUGCGACUGUAAC ...(((..(((((((((.((((..........(....).................((((........))))................)))).)))))))))..)))... ( -28.60) >DroEre_CAF1 128402 91 + 1 AUGGCAAGUGCAGCUUCCGCUGCAACUAAAACGGAAACAGA------------AAUG------AAAAUGGAAGUGCAACUCAAACGGCAGCCGAAGCUGUGACUGUAAC ...(((..(((((((((.(((((.........(....)...------------..((------(...((......))..)))....))))).)))))))))..)))... ( -28.00) >consensus AUGGCAAGUGCAGCUUCCGCUGGAACUAAAACGGAAACAAAAAUGAAAAUGAAAAUGCAACUGAAACUGUAACUGGAACUCAAACGGCAGCCGAAGCUGCGACUGUAAC ...(((..(((((((((.((((..........(....)..................................(((.........))))))).)))))))))..)))... (-25.76 = -25.82 + 0.06)


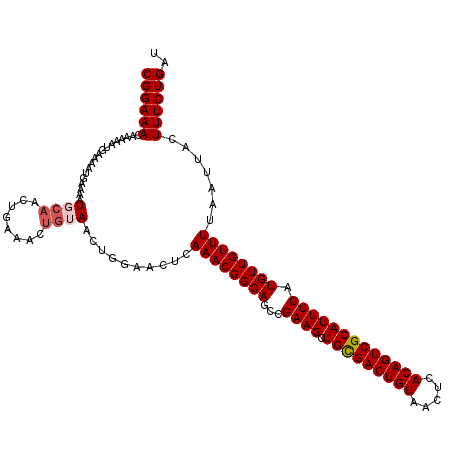
| Location | 13,579,721 – 13,579,838 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 89.17 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -24.91 |
| Energy contribution | -25.72 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13579721 117 + 20766785 CGGAAACAAAAAUGAAAAUGAAAAUGCAACUGAAACUGAAACUGGAACUCAAACGGCAGCCGAAGCUGCGACUGUAACUCACAGUCGCAUUUCAUGUUGUUUUAAUUACUUUCUGAU ((((((..((((..(..((((((.(((............................(((((....)))))((((((.....))))))))))))))).)..))))......)))))).. ( -28.50) >DroSec_CAF1 132777 117 + 1 CGGAAACAAAAAUGAAAAGGAAAAUGCAACUGAAACUGUAACUGGAACUCAAACGGCAGCCGAAGCUGCGACUGUAACUCACAGUCGCAUUUCAUGUUGUUUUAAUUACUUUCUGAU .(....)......((((..((....((((((((((.(((................(((((....)))))((((((.....)))))))))))))).))))).....))..)))).... ( -29.20) >DroSim_CAF1 131155 117 + 1 CGGAAACAAAAAUGAAAAUGAAAAUGCAACUGAAACUGUAACUGGAACUCAAACGGCAGCCGAAGCUGCGACUGUAACUCACAGUCGCAUUUCAUGUUGUUUUAAUUACUUUCUGAU .(....)......((((.(((....((((((((((.(((................(((((....)))))((((((.....)))))))))))))).))))).....))).)))).... ( -30.10) >DroEre_CAF1 128433 99 + 1 CGGAAACAGA------------AAUG------AAAAUGGAAGUGCAACUCAAACGGCAGCCGAAGCUGUGACUGUAACUCACAGUCGCAUUUCAUGUUGUUUUAAUUACUUUCUGAU ((((((((..------------..))------......(((((((..........(((((....)))))((((((.....)))))))))))))................)))))).. ( -28.90) >consensus CGGAAACAAAAAUGAAAAUGAAAAUGCAACUGAAACUGUAACUGGAACUCAAACGGCAGCCGAAGCUGCGACUGUAACUCACAGUCGCAUUUCAUGUUGUUUUAAUUACUUUCUGAU ((((((..................((((........))))..........((((((((...((((.(((((((((.....))))))))))))).)))))))).......)))))).. (-24.91 = -25.72 + 0.81)


| Location | 13,579,721 – 13,579,838 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 89.17 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -22.24 |
| Energy contribution | -23.55 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.842084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13579721 117 - 20766785 AUCAGAAAGUAAUUAAAACAACAUGAAAUGCGACUGUGAGUUACAGUCGCAGCUUCGGCUGCCGUUUGAGUUCCAGUUUCAGUUUCAGUUGCAUUUUCAUUUUCAUUUUUGUUUCCG ....((((((........((((.((((((..(((((.((..(.(((..(((((....)))))...))))..)))))))...)))))))))).......))))))............. ( -31.46) >DroSec_CAF1 132777 117 - 1 AUCAGAAAGUAAUUAAAACAACAUGAAAUGCGACUGUGAGUUACAGUCGCAGCUUCGGCUGCCGUUUGAGUUCCAGUUACAGUUUCAGUUGCAUUUUCCUUUUCAUUUUUGUUUCCG ....(((((.........((((.(((((((..((((.((..(.(((..(((((....)))))...))))..))))))..)).)))))))))........)))))............. ( -30.13) >DroSim_CAF1 131155 117 - 1 AUCAGAAAGUAAUUAAAACAACAUGAAAUGCGACUGUGAGUUACAGUCGCAGCUUCGGCUGCCGUUUGAGUUCCAGUUACAGUUUCAGUUGCAUUUUCAUUUUCAUUUUUGUUUCCG ....((((((........((((.(((((((..((((.((..(.(((..(((((....)))))...))))..))))))..)).))))))))).......))))))............. ( -31.66) >DroEre_CAF1 128433 99 - 1 AUCAGAAAGUAAUUAAAACAACAUGAAAUGCGACUGUGAGUUACAGUCACAGCUUCGGCUGCCGUUUGAGUUGCACUUCCAUUUU------CAUU------------UCUGUUUCCG ..((((((((((((.((((............(((((((...))))))).((((....))))..)))).))))))...........------..))------------))))...... ( -25.01) >consensus AUCAGAAAGUAAUUAAAACAACAUGAAAUGCGACUGUGAGUUACAGUCGCAGCUUCGGCUGCCGUUUGAGUUCCAGUUACAGUUUCAGUUGCAUUUUCAUUUUCAUUUUUGUUUCCG ..((((((((.......)).....((((((((((((((...)))))))(((((....)))))............................)))))))........))))))...... (-22.24 = -23.55 + 1.31)

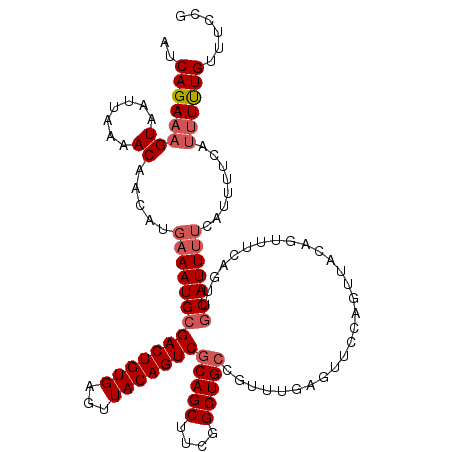

| Location | 13,579,760 – 13,579,861 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.20 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -19.50 |
| Energy contribution | -20.78 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13579760 101 + 20766785 AACUGGAACUCAAACGGCAGCCGAAGCUGCGACUGUAACUCACAGUCGCAUUUCAUGUUGUUUUAAUUACUUUCUGAUUUAUUUAGAGUGGUGCCAAGG---GG ........(((((((((((...((((.(((((((((.....))))))))))))).))))))))....((((((((((.....)))))).))))....))---). ( -31.00) >DroSec_CAF1 132816 101 + 1 AACUGGAACUCAAACGGCAGCCGAAGCUGCGACUGUAACUCACAGUCGCAUUUCAUGUUGUUUUAAUUACUUUCUGAUUUAUUUAGAAUGGUGUCAAGG---GG ........(((((((((((...((((.(((((((((.....))))))))))))).))))))))....((((((((((.....)))))).))))....))---). ( -31.50) >DroSim_CAF1 131194 101 + 1 AACUGGAACUCAAACGGCAGCCGAAGCUGCGACUGUAACUCACAGUCGCAUUUCAUGUUGUUUUAAUUACUUUCUGAUUUAUUUAGAAUGGUGCCAAGG---GG ........(((((((((((...((((.(((((((((.....))))))))))))).))))))))....((((((((((.....)))))).))))....))---). ( -31.40) >DroEre_CAF1 128454 101 + 1 AAGUGCAACUCAAACGGCAGCCGAAGCUGUGACUGUAACUCACAGUCGCAUUUCAUGUUGUUUUAAUUACUUUCUGAUUUAUUUAGAGUGGUGCCAAGG---AG ........(((....((((.((((((.(((((((((.....))))))))))))).................((((((.....)))))).))))))...)---)) ( -29.40) >DroYak_CAF1 132623 101 + 1 AAGUGCGACUCCAACGACAGCCGAAGCUGUGACUGUAACUCACAGUCGCAUUUCAUGUUGUUUUAAUUACUUUCUGAUUUAUUUAGAGUGGUGGCAAGG---GG ...(((.((..((((((((...((((.(((((((((.....))))))))))))).)))))))..........(((((.....))))))..)).)))...---.. ( -31.80) >DroAna_CAF1 125367 88 + 1 ----------------GGGGCCAAAACUGCGACCGUAAUUCACUAACAAAUUUCGUGUUGUUUUAAUUACUUUCUGAUUUAUUUAACGGAGCGACUCGGUGGUC ----------------(.((......)).)((((((((((....(((((........)))))..))))))...............((.(((...))).)))))) ( -13.70) >consensus AACUGGAACUCAAACGGCAGCCGAAGCUGCGACUGUAACUCACAGUCGCAUUUCAUGUUGUUUUAAUUACUUUCUGAUUUAUUUAGAGUGGUGCCAAGG___GG ...............((((.((((((.(((((((((.....))))))))))))).................((((((.....)))))).))))))......... (-19.50 = -20.78 + 1.28)


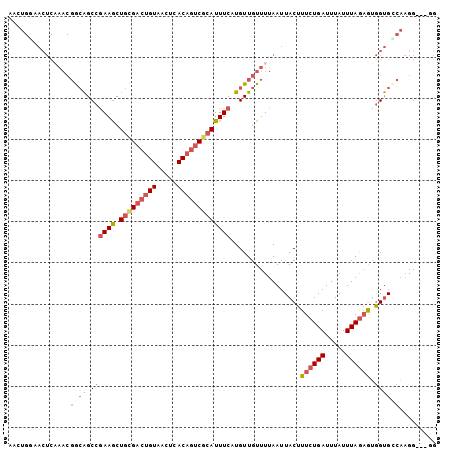
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:38 2006