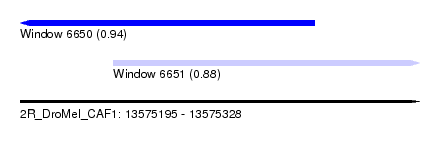
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,575,195 – 13,575,328 |
| Length | 133 |
| Max. P | 0.940060 |

| Location | 13,575,195 – 13,575,293 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 73.59 |
| Mean single sequence MFE | -30.76 |
| Consensus MFE | -15.82 |
| Energy contribution | -17.32 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
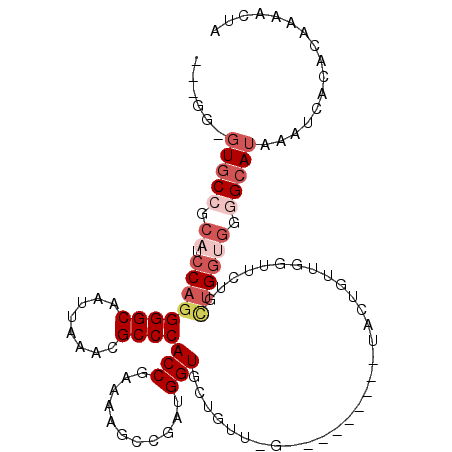
>2R_DroMel_CAF1 13575195 98 - 20766785 ---GG-GUGCCGCAUCCAGGGGCAAUUAAACGCCCACCGAAAAGCCGAUGGUGCUGUUCG---------UACUGUUGGUUCUGCUGGUGGGGCAUAAAUCACACAAAACUA ---..-(((((.((.((((((((........)))).......((((((((((((.....)---------)))))))))))...)))))).)))))................ ( -38.70) >DroSec_CAF1 122165 93 - 1 ---GG-GUGCCGCAUCCACGGGCAAUUAAGCGCCCACCGAAAAGCCGAUGGUACUGUUGG---------UACUGUUCGUUCUGCUG-----GCAUAAAUCACACAAAACUA ---..-((((((((.....((((........))))..((((..((((((......)))))---------)....))))...))).)-----))))................ ( -26.90) >DroSim_CAF1 126670 89 - 1 ---GG-GUGCCGCAUCCAGGGGCAAUUAAACGCCCACCGAAAAGUGGAUGGUAC------------------UGUUCGUUCUGCUGGUGGGGCAUAAAUCACACAAAACUA ---..-(((((.((.((((((((........))))..((((.((((.....)))------------------).)))).....)))))).)))))................ ( -29.90) >DroEre_CAF1 124034 80 - 1 ---GG-GUGCCGCAUCCAGGGGCAAUUAAACGCCCACCGAAAUGCCGAUGGUGCU---------------------------GUUGGUGGGGCAUAAAUCACACAAAACUA ---..-(((((.((.((((.((((.........(((.((......)).)))))))---------------------------.)))))).)))))................ ( -24.50) >DroYak_CAF1 127911 107 - 1 ---GG-GUGCCGCAUCCAGGGGCAAUUAAACGCCCACCGAAAUACCGAUGGUGCUGUUGGUGAUGAUGGUGCUGUUGGUGCUGUUGGUGGGGCAUAAAUCACACAAAACUA ---..-(((((.(((((((((((........)))).......(((((((((..(((((......)))))..))))))))))))..)))).)))))................ ( -40.80) >DroAna_CAF1 121392 92 - 1 AUAAACUUCCAACAUCCAGGGGCAAUCAAACGCCCACCGGAUUUC----GGUGCGGUACA---------CACA--UGCC----UCGGCGGAGCAUAAAUCACACAAAACUA ..............(((.((((((.......((((((((.....)----)))).)))...---------....--))))----))...))).................... ( -23.74) >consensus ___GG_GUGCCGCAUCCAGGGGCAAUUAAACGCCCACCGAAAAGCCGAUGGUGCUGUU_G_________UACUGUUGGUUCUGCUGGUGGGGCAUAAAUCACACAAAACUA ......(((((.((.((((((((........))))(((...........)))...............................)))))).)))))................ (-15.82 = -17.32 + 1.50)

| Location | 13,575,226 – 13,575,328 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.02 |
| Mean single sequence MFE | -37.68 |
| Consensus MFE | -22.32 |
| Energy contribution | -23.27 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.878794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
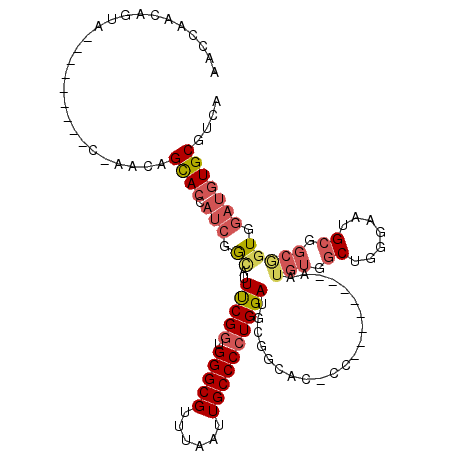
>2R_DroMel_CAF1 13575226 102 + 20766785 AACCAACAGUA---------CGAACAGCACCAUCGGCUUUUCGGUGGGCGUUUAAUUGCCCCUGGAUGCGGCAC-CC--------AAUGUGGCUGGGAAUGCGGCAGUGGAUGUGCGUCA ...........---------.((...((((.(((.((..(((((.(((((......))))))))))(((.((((-((--------(.......))))..))).))))).))))))).)). ( -39.20) >DroSec_CAF1 122191 102 + 1 AACGAACAGUA---------CCAACAGUACCAUCGGCUUUUCGGUGGGCGCUUAAUUGCCCGUGGAUGCGGCAC-CC--------AAUGUGGCUGGGAAUGCGGCGGUGGAUGUGCGUCA .(((.(((.((---------((.......((((.(((..((.(((....))).))..))).))))..((.((((-((--------(.......))))..))).))))))..))).))).. ( -34.60) >DroSim_CAF1 126701 93 + 1 AACGAACA------------------GUACCAUCCACUUUUCGGUGGGCGUUUAAUUGCCCCUGGAUGCGGCAC-CC--------AAUGUGGCUGGGAAUGCGGCGGUGGAUGUGCGUCA ...((...------------------((((.(((((((.(((((.(((((......)))))))))).((.((((-((--------(.......))))..))).))))))))))))).)). ( -40.90) >DroEre_CAF1 124063 86 + 1 -------------------------AGCACCAUCGGCAUUUCGGUGGGCGUUUAAUUGCCCCUGGAUGCGGCAC-CC--------AAUGUGGCUGGGAAUGCGGCGGUGGAUGUGCGUCA -------------------------.((((.(((.((..(((((.(((((......))))))))))(((.((((-((--------(.......))))..))).))))).))))))).... ( -37.50) >DroYak_CAF1 127942 111 + 1 CACCAACAGCACCAUCAUCACCAACAGCACCAUCGGUAUUUCGGUGGGCGUUUAAUUGCCCCUGGAUGCGGCAC-CC--------AAUGUGGCUGGGAAUGCGGCGGUGGAUGUGCGUCA ........((((.(((..((((.................(((((.(((((......)))))))))).((.((((-((--------(.......))))..))).))))))))))))).... ( -38.80) >DroAna_CAF1 121420 98 + 1 -GGCA--UGUG---------UGUACCGCACC----GAAAUCCGGUGGGCGUUUGAUUGCCCCUGGAUGUUGGAAGUUUAUAUAUAAGCAUGGCUGGGAAUG------UGAAUUUGCGUCA -.((.--((((---------((((.....((----((.((((((.(((((......))))))))))).)))).....)))))))).)).((((.(.((((.------...)))).))))) ( -35.10) >consensus AACCAACAGUA_________C_AACAGCACCAUCGGCAUUUCGGUGGGCGUUUAAUUGCCCCUGGAUGCGGCAC_CC________AAUGUGGCUGGGAAUGCGGCGGUGGAUGUGCGUCA ..........................((((.(((.((..(((((.(((((......)))))))))).....................(((.((.......)).))))).))))))).... (-22.32 = -23.27 + 0.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:33 2006