| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,571,945 – 13,572,037 |
| Length | 92 |
| Max. P | 0.996402 |

| Location | 13,571,945 – 13,572,037 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.53 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -12.69 |
| Energy contribution | -13.30 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.42 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
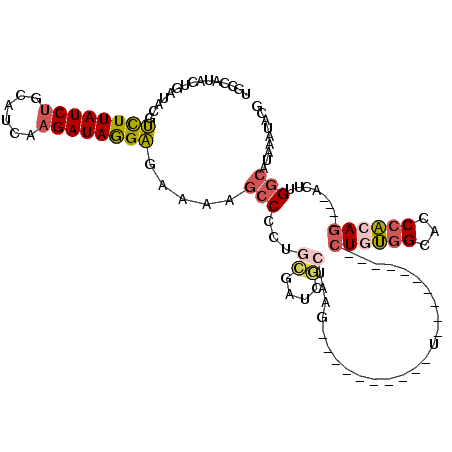
>2R_DroMel_CAF1 13571945 92 - 20766785 UGGCAUACUGAUAUGUCUUAUCUGCAUCAAGAUAGGAGAAAAGCCCCUGCGAUCGCUAAG----------U----------CUGUGGCACCCAAAG----ACUUGGCAUAAAUGCC .(((((...(((.(.((((((((......)))))))).)...((....)).)))((((((----------(----------((.(((...))).))----)))))))....))))) ( -32.70) >DroSec_CAF1 118717 92 - 1 UGCCGUACUGAUACGUCUUAUCUGCAUCAAGAUAGGAGAAAAGCCCCAGCGAUCGCUAAG----------U----------CUGUGGCACCCACAG----ACUUGGCAUAAAUACU .((((((....))))((((((((......))))))))...........))....((((((----------(----------((((((...))))))----)))))))......... ( -33.50) >DroSim_CAF1 123217 92 - 1 UGCCGUACUGAUACGUCUUAUCUGCAUCAAGAUAGGAGAAAAGCCCCAGCGAUCGCUAAG----------U----------CUGUGGCACCCACAG----ACUUGGCAUAAAUACG ...((((..(((.(.((((((((......)))))))).....((....))))))((((((----------(----------((((((...))))))----))))))).....)))) ( -34.50) >DroEre_CAF1 120607 92 - 1 UGUCACACUGAUACGUCUUAUCUGCAUUAAGAUAGGAGGAAAGCCCCUCCGAUUCCUAAG----------C----------CUAUGGCACCCACAG----AGUAGGCAUAAAUACG ((((((.(((....(((.(((((......)))))(((((......))))))))......(----------(----------(...))).....)))----.)).))))........ ( -23.50) >DroYak_CAF1 124554 92 - 1 UGUCAUACUGAUACGUCUUAUCUGCACUAAGAUAGGAGAAAAGCCCCUGCGAUCGCUAAG----------U----------CUGUGGCACCCACAG----AGUUGGCAUAAAUACG ((((.....))))..((((((((......))))))))((...((....))..))(((((.----------(----------((((((...))))))----).)))))......... ( -28.80) >DroPer_CAF1 124903 111 - 1 UCCAUUAGUGAUUGUCUUUAUCAGUGGCAAGAUAAGGGG-----CCCCUGAGUCCCAGUGGUCUUGUAAAUUCAUAGAUAAUUGGUGUUCCCUCAUUGUUACGUGCCAUAAAAGCA ........((((.......))))((((((.(((((((((-----.....(((..(((((.((((((......)).)))).)))))..))))))).)))))...))))))....... ( -27.00) >consensus UGCCAUACUGAUACGUCUUAUCUGCAUCAAGAUAGGAGAAAAGCCCCUGCGAUCGCUAAG__________U__________CUGUGGCACCCACAG____ACUUGGCAUAAAUACG ...............((((((((......)))))))).....(((...((....)).........................((((((...))))))........)))......... (-12.69 = -13.30 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:31 2006