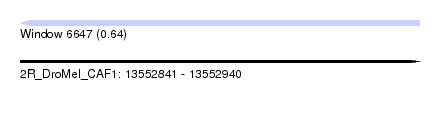
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,552,841 – 13,552,940 |
| Length | 99 |
| Max. P | 0.637275 |

| Location | 13,552,841 – 13,552,940 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.99 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -19.02 |
| Energy contribution | -18.38 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
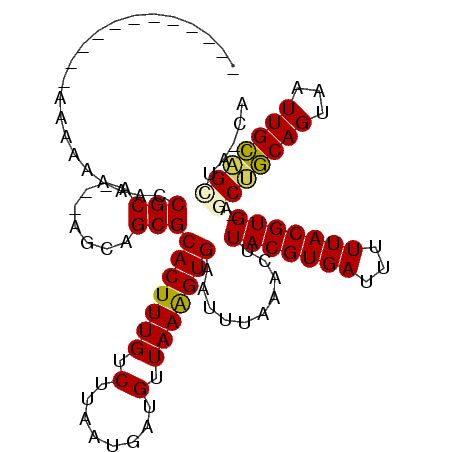
>2R_DroMel_CAF1 13552841 99 - 20766785 ------------AAAAAGAACCGCAGCAAUGUGCGCACUUUGUCUUAAUGAUGUUAAAGUGAAUUUAAACUUACGUGAUUUUUACGUG-AGCUGCAGUAAUUGCCGAUGCCA ------------..........((((((((.(((.(((((((.(........).)))))))........(((((((((...)))))))-))..)))...)))))...))).. ( -24.00) >DroVir_CAF1 90467 96 - 1 ------------AUGAAAAACCGCA---AGCAGCGCACUUUGUCUUAAUGAUGUUAAGGUGAAUUUAAACUUACGUGAUUUUUACGUGCAGCUGCAGUAAUUGCAGUAA-UG ------------.......((.(((---((((((((((((..(((((((...)))))))..))...........((((...)))))))).))))).....)))).))..-.. ( -24.20) >DroPse_CAF1 99491 109 - 1 UCUCCUAAGUCGAAACAGAACCGCAGU-GUGUGCGCACUUUGUCUUAAUGAUGUUAAAGUGAAUUUAAACUUACGUGAUUUUUACGUG-AGCGCCAGUAAUUGGCGCUG-CA ......................(((((-((.(((.(((((((.(........).)))))))........(((((((((...)))))))-)).....)))....))))))-). ( -27.20) >DroGri_CAF1 77738 96 - 1 ------------GGCAAAAACCGCA---AGCAGCGCACUUUGUCUUAAUGAUGUUAAGGUGAAUUUCAACUUACGUGAUUUUUACGUGCAACUGCAGUAAUUGCAGCUA-UG ------------((......))(((---(((((.((((.....((((((...))))))(((((..(((.......)))..)))))))))..)))).....)))).....-.. ( -21.70) >DroMoj_CAF1 114966 96 - 1 ------------GGGAAAAACCGCA---AGCAGCGCACUUUGUCUUAAUGAUGUUAAGGUGAAUUUAAACUUACGUGAUUUUUACGUGCAGCUGCAGUAAUUGCAGUAA-UG ------------((......))(((---((((((((((((..(((((((...)))))))..))...........((((...)))))))).))))).....)))).....-.. ( -25.90) >DroPer_CAF1 107536 109 - 1 UCUUCUAAGUCGAAACAGAACCGCAGU-GUGUGCGCACUUUGUCUUAAUGAUGUUAAAGUGAAUUUAAACUUACGUGAUUUUUACGUG-AGCGCCAGUAAUUGGCGCUG-CA ......................(((((-((.(((.(((((((.(........).)))))))........(((((((((...)))))))-)).....)))....))))))-). ( -27.20) >consensus ____________AAAAAAAACCGCA___AGCAGCGCACUUUGUCUUAAUGAUGUUAAAGUGAAUUUAAACUUACGUGAUUUUUACGUG_AGCUGCAGUAAUUGCAGCUA_CA .....................(((........)))(((((((.(........).)))))))..........(((((((...)))))))..(((((((...)))))))..... (-19.02 = -18.38 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:29 2006