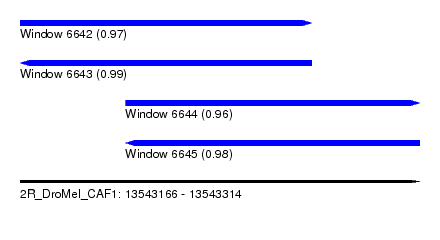
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,543,166 – 13,543,314 |
| Length | 148 |
| Max. P | 0.994619 |

| Location | 13,543,166 – 13,543,274 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -50.83 |
| Consensus MFE | -45.41 |
| Energy contribution | -45.30 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
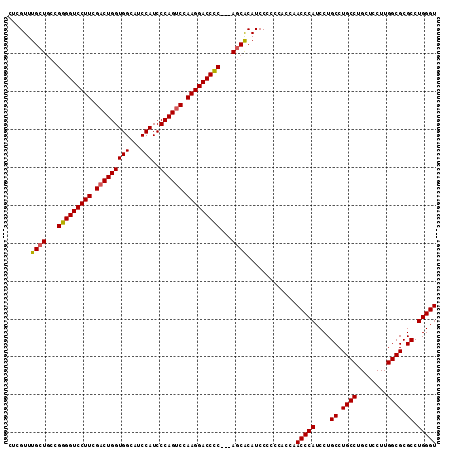
>2R_DroMel_CAF1 13543166 108 + 20766785 ACCCAGGCGCGCCAAGGAGCUGGCAGGCAGGAUGGGUUU-----GGGAUGCGCUGAGGAGGUCCUUGGACUGGGAUGGAUGCCACCAGUCGAAGGACCCCAGGAGCAAACGAG .(((((((.((((.....(((....))).)).)).))))-----))).(((.((..((.(((((((.((((((..(((...))))))))).))))))))).)).)))...... ( -48.70) >DroSec_CAF1 90852 110 + 1 ACCCAGGCGCGCCAAGGAGCAGGCAGGCAGGAUGGGUUGGUGGGGGGAUGUGAU---GGGGUCCUUGGACUGGGAUGGAUGCCACCAGUCGAAGGACCCCGGCAGCAAACGAG (((((.((..(((........)))..))....)))))........(..(((..(---(((((((((.((((((..(((...))))))))).))))))))))...)))..)... ( -49.70) >DroSim_CAF1 94808 110 + 1 ACCCAGGCGCGCCAAGGAGCAGGCAGGCAGGAUGGGUUGGUGGGGGGAUGUGCU---GGGGUCCUUGGGCUGGGAUGGAUGCCACCAGUCGAAGGACCCCGGCAGCAAACGAG (((((.((..(((........)))..))....)))))........(..((((((---(((((((((.((((((..(((...))))))))).))))))))))))).))..)... ( -54.10) >consensus ACCCAGGCGCGCCAAGGAGCAGGCAGGCAGGAUGGGUUGGUGGGGGGAUGUGCU___GGGGUCCUUGGACUGGGAUGGAUGCCACCAGUCGAAGGACCCCGGCAGCAAACGAG (((((.((..(((........)))..))....)))))........(..(((......(((((((((.((((((..(((...))))))))).)))))))))....)))..)... (-45.41 = -45.30 + -0.11)

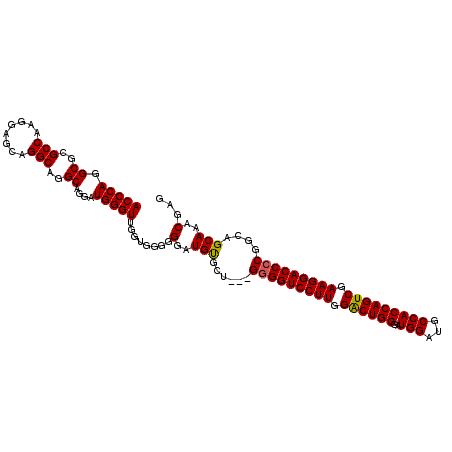

| Location | 13,543,166 – 13,543,274 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -43.93 |
| Consensus MFE | -42.37 |
| Energy contribution | -42.60 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13543166 108 - 20766785 CUCGUUUGCUCCUGGGGUCCUUCGACUGGUGGCAUCCAUCCCAGUCCAAGGACCUCCUCAGCGCAUCCC-----AAACCCAUCCUGCCUGCCAGCUCCUUGGCGCGCCUGGGU .......(((...(((((((((.(((((((((...)))..)))))).)))))))))...))).......-----..(((((....((.((((((....)))))).)).))))) ( -44.10) >DroSec_CAF1 90852 110 - 1 CUCGUUUGCUGCCGGGGUCCUUCGACUGGUGGCAUCCAUCCCAGUCCAAGGACCCC---AUCACAUCCCCCCACCAACCCAUCCUGCCUGCCUGCUCCUUGGCGCGCCUGGGU ...(..((.((..(((((((((.(((((((((...)))..)))))).)))))))))---..))))..)........(((((....((.((((........)))).)).))))) ( -44.10) >DroSim_CAF1 94808 110 - 1 CUCGUUUGCUGCCGGGGUCCUUCGACUGGUGGCAUCCAUCCCAGCCCAAGGACCCC---AGCACAUCCCCCCACCAACCCAUCCUGCCUGCCUGCUCCUUGGCGCGCCUGGGU ...(..((.(((.(((((((((.(.(((((((...)))..)))).).)))))))))---.)))))..)........(((((....((.((((........)))).)).))))) ( -43.60) >consensus CUCGUUUGCUGCCGGGGUCCUUCGACUGGUGGCAUCCAUCCCAGUCCAAGGACCCC___AGCACAUCCCCCCACCAACCCAUCCUGCCUGCCUGCUCCUUGGCGCGCCUGGGU ......((((...(((((((((.(((((((((...)))..)))))).)))))))))...)))).............(((((....((.((((........)))).)).))))) (-42.37 = -42.60 + 0.23)

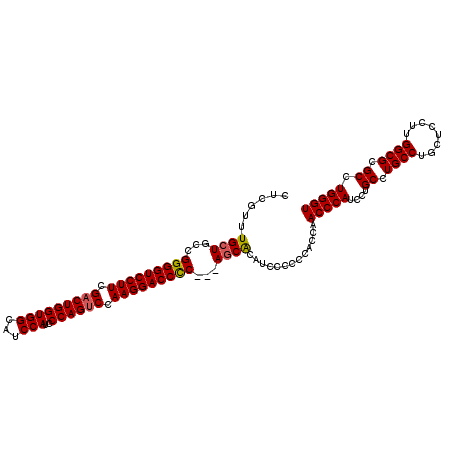
| Location | 13,543,205 – 13,543,314 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 89.68 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -35.67 |
| Energy contribution | -36.23 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.67 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
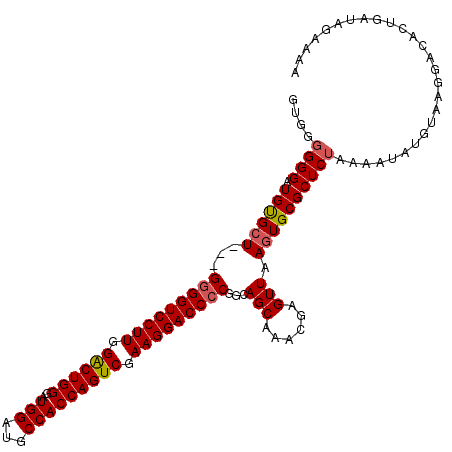
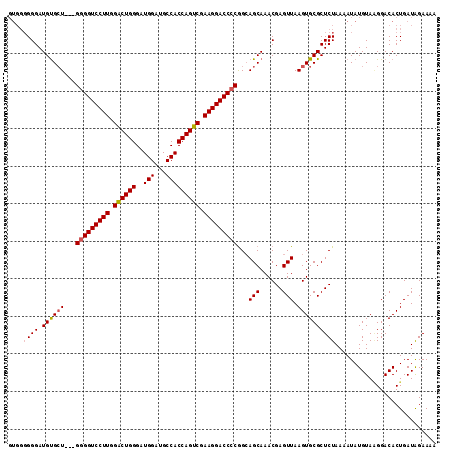
>2R_DroMel_CAF1 13543205 109 + 20766785 -----GGGAUGCGCUGAGGAGGUCCUUGGACUGGGAUGGAUGCCACCAGUCGAAGGACCCCAGGAGCAAACGAGUUAAGUGCGCUCUAAAAUAUGUAGGGACACUGAUGGAAAA -----.(..(((.((..((.(((((((.((((((..(((...))))))))).))))))))).)).)))..)......((((..(((((.......))))).))))......... ( -40.70) >DroSec_CAF1 90891 111 + 1 GUGGGGGGAUGUGAU---GGGGUCCUUGGACUGGGAUGGAUGCCACCAGUCGAAGGACCCCGGCAGCAAACGAGUUAAGUGCGCUCUAAUAUAUGUACGGACACUGAUAGAAAA (((...(..(((..(---(((((((((.((((((..(((...))))))))).))))))))))...)))..)((((.......))))...............))).......... ( -41.80) >DroSim_CAF1 94847 111 + 1 GUGGGGGGAUGUGCU---GGGGUCCUUGGGCUGGGAUGGAUGCCACCAGUCGAAGGACCCCGGCAGCAAACGAGUUAAGUGCGCUCUAAAAUAUGUAAGGACACUGAUAGAAAA (((...(..((((((---(((((((((.((((((..(((...))))))))).))))))))))))).))..)((((.......))))...............))).......... ( -46.20) >consensus GUGGGGGGAUGUGCU___GGGGUCCUUGGACUGGGAUGGAUGCCACCAGUCGAAGGACCCCGGCAGCAAACGAGUUAAGUGCGCUCUAAAAUAUGUAAGGACACUGAUAGAAAA ....((((.((((((...(((((((((.((((((..(((...))))))))).)))))))))...(((......))).))))))))))........................... (-35.67 = -36.23 + 0.56)

| Location | 13,543,205 – 13,543,314 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 89.68 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -32.11 |
| Energy contribution | -32.33 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.06 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
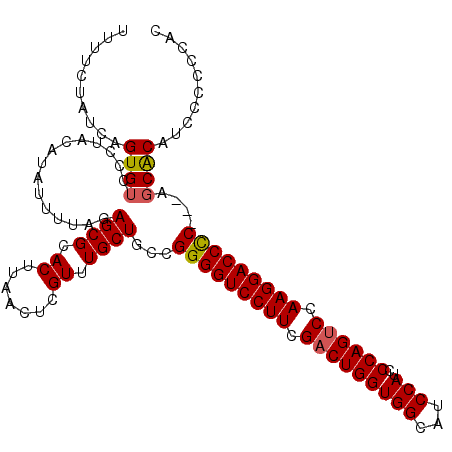
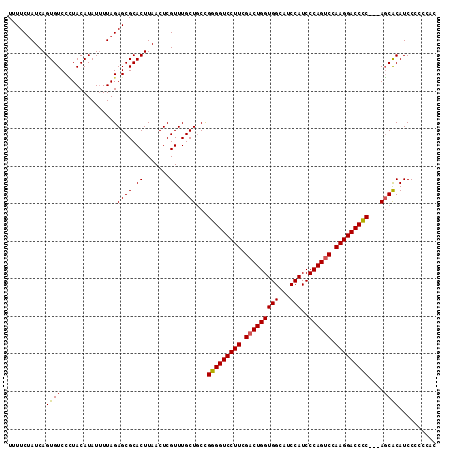
>2R_DroMel_CAF1 13543205 109 - 20766785 UUUUCCAUCAGUGUCCCUACAUAUUUUAGAGCGCACUUAACUCGUUUGCUCCUGGGGUCCUUCGACUGGUGGCAUCCAUCCCAGUCCAAGGACCUCCUCAGCGCAUCCC----- ..........((((..............(((((.((.......)).)))))..(((((((((.(((((((((...)))..)))))).)))))))))....)))).....----- ( -35.90) >DroSec_CAF1 90891 111 - 1 UUUUCUAUCAGUGUCCGUACAUAUAUUAGAGCGCACUUAACUCGUUUGCUGCCGGGGUCCUUCGACUGGUGGCAUCCAUCCCAGUCCAAGGACCCC---AUCACAUCCCCCCAC ..........(((................((((.((.......)).))))...(((((((((.(((((((((...)))..)))))).)))))))))---..))).......... ( -36.00) >DroSim_CAF1 94847 111 - 1 UUUUCUAUCAGUGUCCUUACAUAUUUUAGAGCGCACUUAACUCGUUUGCUGCCGGGGUCCUUCGACUGGUGGCAUCCAUCCCAGCCCAAGGACCCC---AGCACAUCCCCCCAC .........(((((.(((..........))).)))))......(..((.(((.(((((((((.(.(((((((...)))..)))).).)))))))))---.)))))..)...... ( -35.20) >consensus UUUUCUAUCAGUGUCCCUACAUAUUUUAGAGCGCACUUAACUCGUUUGCUGCCGGGGUCCUUCGACUGGUGGCAUCCAUCCCAGUCCAAGGACCCC___AGCACAUCCCCCCAC ..........((((...............((((.((.......)).))))...(((((((((.(((((((((...)))..)))))).)))))))))....)))).......... (-32.11 = -32.33 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:27 2006