
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,536,517 – 13,536,671 |
| Length | 154 |
| Max. P | 0.723555 |

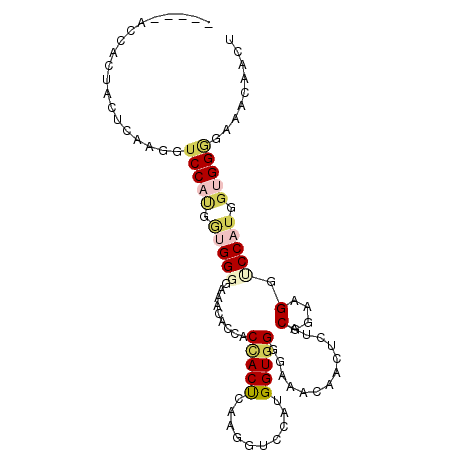
| Location | 13,536,517 – 13,536,611 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 71.44 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -12.07 |
| Energy contribution | -13.02 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13536517 94 + 20766785 -----ACUACUACUCAAGGUCCAUGGUGGGGAAACACUACCACUCAAGGUCCAUGGUGGGGAAACACGACUACUCAAGGUCCAUGGUGGGGAAACACCA -----....((((.((.((.((.(((((((....).)))))).....)).)).))))))(....)..((((......))))..(((((......))))) ( -31.10) >DroSim_CAF1 88118 99 + 1 GAAACACUACUACUCAAGGUCCAUGGUGGGGAAACAGCACUACUCAAGGUCCAUGGUGGGGAAACAACUCAACGGAAGGUCCAUGGUGGGGAAACAACU ....(((((((((.((.((.((.(((((((....)....)))).)).)).)).))))))(((..........(....).))).))))).(....).... ( -26.20) >DroEre_CAF1 85351 94 + 1 -----ACCACUACUCAAGGUCCAUGGUGGGGAAACACCACUACUCAAGGUCCAUGGUGGGGAAACACCACUACUCAAGGCCCAUGGUGGGGAAACUCUA -----.((.((((.((.((.((.(((((((....).))))))...........(((((((....).)))))).....)).)).))))))))........ ( -34.30) >DroWil_CAF1 65040 85 + 1 -----UCCACGGCAUAUCCUCCCCAAUGG---AACACCACCACCGAUCGUCCUUGGUGG---AACAAUUCUACCGAAGGUCCUUGGUGGAAU---AACU -----.....((....(((........))---)...)).(((((((..(.(((((((((---(.....)))))).)))).).)))))))...---.... ( -27.50) >DroYak_CAF1 88290 94 + 1 -----AACUCUACUCAAGGUCCAUGGUGGGGAAACAACUCAACUGAAGGUCCCUGGUGGGGAAACAACUCGACGGAAGGUCCAUGGUGGGGAAACAACU -----..((((((.((.((.((.....(((((.....(((....).)).))))).((.(((......))).))....)).)).))))))))........ ( -25.70) >DroAna_CAF1 86093 88 + 1 -----AAUUCUACCUCUGGACCCAACUGG------AACACCACUGAAGGACCGUGGUGGGGAAAUAACUCGACGGAAGGACCCUGGUGGGGCAACAACU -----....(((((...((..((..(((.------..((((((.(.....).))))))(((......)))..)))..))..)).)))))(....).... ( -25.40) >consensus _____ACCACUACUCAAGGUCCAUGGUGGGGAAACACCACCACUCAAGGUCCAUGGUGGGGAAACAACUCUACGGAAGGUCCAUGGUGGGGAAACAACU ...................(((((.(((((.........(((((..........))))).............(....).))))).)))))......... (-12.07 = -13.02 + 0.95)


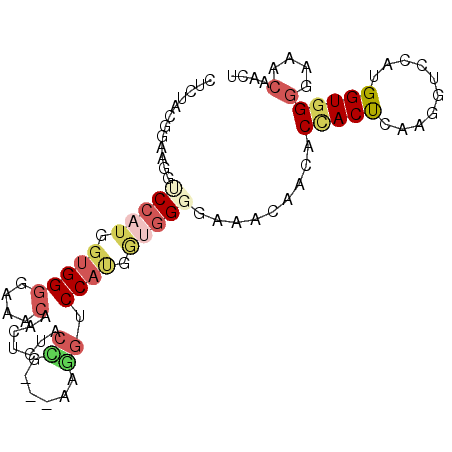
| Location | 13,536,579 – 13,536,671 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 70.69 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -10.89 |
| Energy contribution | -12.03 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13536579 92 + 20766785 GACUACUCAAGGUCCAUGGUGGGGAAACACCACUACUC---AAGGCCCAUGGUGGGGAAACACUACCACUCAAGGUCCAUGGUGGGGAAACACAA ..((((.((.((.((.(((((((....).))))))...---........(((((((....).)))))).....)).)).))))))(....).... ( -34.30) >DroSim_CAF1 88185 92 + 1 CUCAACGGAAGGUCCAUGGUGGGGAAACAACUCAACGG---AAGGUCCAUGGUGGGGCAAUAACUCUACUCAAGGCCCAUGGUGGGGAAACACCA ..........(((((((.(((((............(..---..)..((.((((((((......)))))).)).))))))).))))(....)))). ( -30.10) >DroEre_CAF1 85413 92 + 1 CACUACUCAAGGCCCAUGGUGGGGAAACUCUACUACUC---AAGGUCCAUGGUGGGGAAACACUACUACUCAAGGUCCAUGGUGGGGAAACAACU ..((((.((.((.((.(((((((....).))))))...---........(((((((....).)))))).....)).)).))))))(....).... ( -28.60) >DroWil_CAF1 65096 86 + 1 UUCUACCGAAGGUCCUUGGUGGAAU---AACUCUACGGCAUCUCCUCCACAAUUG---AACACCACCACCGAUCGCCCCUGGUGG---AACAAUU ((((((((..(((..(((((((...---........((........)).....((---.....)))))))))..)))..))))))---))..... ( -23.80) >DroYak_CAF1 88352 92 + 1 CUCGACGGAAGGUCCAUGGUGGGGAAACAACUCAACCG---AAGGUCCAUGGUGGGGAAACAACUCAACUCAAGGUCCAUGGUGGGGAAACAACU .....(....)..((((.(((((....)......(((.---..(((....(((..(....).)))..)))...))))))).))))(....).... ( -23.20) >DroAna_CAF1 86149 86 + 1 CUCGACGGAAGGACCCUGGUGGGGCAACAACUCUACCU---CUGGACCCAACUGG------AACACCACCGAAGGACCAUGGUGGGGAAACAAUU .....(((..((..((.(((((((......))))))).---..))..))..))).------....((((((........))))))(....).... ( -26.90) >consensus CUCUACGGAAGGUCCAUGGUGGGGAAACAACUCUACCG___AAGGUCCAUGGUGGGGAAACAACACCACUCAAGGUCCAUGGUGGGGAAACAACU ............(((((.(((((....).......((......)).)))).))))).........(((((..........)))))(....).... (-10.89 = -12.03 + 1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:20 2006