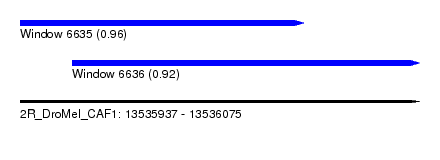
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,535,937 – 13,536,075 |
| Length | 138 |
| Max. P | 0.957036 |

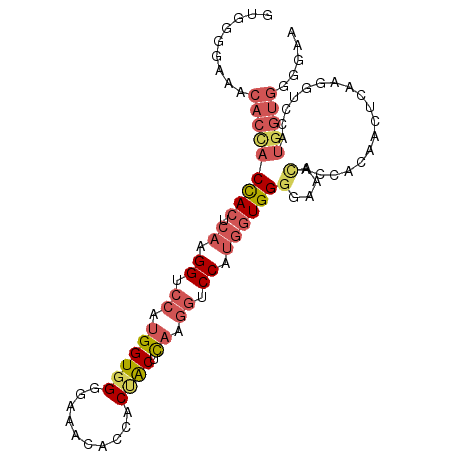
| Location | 13,535,937 – 13,536,035 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 74.59 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -14.94 |
| Energy contribution | -16.62 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.957036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13535937 98 + 20766785 GUGGGGAAACACCACUACUCAAGGUCCAUGGUGGGGAAAUACUACUACUCAAGGUCCAUGGUGGGGAAACACUACCACUCAAGGUCCAUGGUGGGGAA (((((....).)))).........(((((.(((((((....((........)).))).(((((((....).))))))........)))).)))))... ( -33.80) >DroSim_CAF1 87116 98 + 1 GUGGGGAAACACUACCACUCAAGGCCCAUGGUGGGGAAACACAACUACUCAAGGUCCAUGGUGGGGAAACACCACUACUGAAGGUCCAUGGUGGGGAA (((((....).)))).........(((((.(((((....)..............((..(((((((....).))))))..))....)))).)))))... ( -33.60) >DroEre_CAF1 84771 98 + 1 GUUGGGAAACACCACUACUCAAGGUCCAUGGUGGGGAAAUAACUCUACUCAAGGUCCAUGGUGGGGAAAUAACUCUACUCAAGGUCCAUGGUGGGGAA ....(....).((((((...........))))))........((((((.((.((.((.((((((((......)))))).)).)).)).)))))))).. ( -30.90) >DroWil_CAF1 64124 86 + 1 UUGG---AA---CACCACCGAAGGUCCUUGGUGGAAUAACUCCACGGCCUCCCCUCCCCAAUGG---AACACCACAGCAUUACCUCCCCAAUUG---A ((((---..---..(((((((......)))))))......((((.((..........))..)))---)...................))))...---. ( -16.10) >DroYak_CAF1 87710 98 + 1 GUGGGGAAACACCACCACUCAAGGUCCAUGGUGGGGAAACACCACCACUCAAGGUCCAUGGUGGGGAAACAACUCAACGGAAGGUCCAUGGUGGGGAA (((((....).)))).........(((.(((((((....).))))))........((((.(((((....).......(....)..)))).))))))). ( -38.70) >consensus GUGGGGAAACACCACCACUCAAGGUCCAUGGUGGGGAAACACCACUACUCAAGGUCCAUGGUGGGGAAACACCACAACUCAAGGUCCAUGGUGGGGAA .........(((((((((.((.((.((.((((((..........)))).)).)).)).))))))(....)..................)))))..... (-14.94 = -16.62 + 1.68)

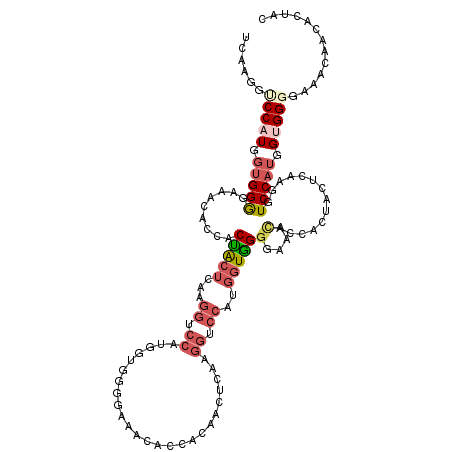
| Location | 13,535,955 – 13,536,075 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.47 |
| Mean single sequence MFE | -37.29 |
| Consensus MFE | -13.72 |
| Energy contribution | -14.81 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.37 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13535955 120 + 20766785 UCAAGGUCCAUGGUGGGGAAAUACUACUACUCAAGGUCCAUGGUGGGGAAACACUACCACUCAAGGUCCAUGGUGGGGAAACACCACUACUCAAGGUCCAUGGUGGGGAAACACCACUAC ....((.((.((((((((........((((.((.((.((.(((((((....).)))))).....)).)).))))))(....).)).)))).)).)).)).(((((((....).)))))). ( -46.80) >DroPse_CAF1 81255 102 + 1 UGAAUGGCCCUGGUGGAGCAAUACGACCGCU---GGACCAA-AUGUGAAUA-----CAACCCAAGGUCCUUGGUGGGGUAACACCA---CUUCCGGUCCAA-GUGGG-----AACACUAC ....(((((..(((((.(...(((..(((((---(((((..-..((.....-----..))....)))))..))))).))).).)))---))...)).)))(-(((..-----..)))).. ( -33.00) >DroSim_CAF1 87134 120 + 1 UCAAGGCCCAUGGUGGGGAAACACAACUACUCAAGGUCCAUGGUGGGGAAACACCACUACUGAAGGUCCAUGGUGGGGAAACACUACGACUCAAGGUCCAUGGUGGGGAAACACAACUAC ......(((((.(((((....)...(((...((.((.((.(((((((....).)))))).....)).)).))(((((....).)))).......))))))).)))))............. ( -38.50) >DroEre_CAF1 84789 120 + 1 UCAAGGUCCAUGGUGGGGAAAUAACUCUACUCAAGGUCCAUGGUGGGGAAAUAACUCUACUCAAGGUCCAUGGUGGGGAAACACCACUACUCAAGGUCCAUGGUGGGGAAAUAACUCCAC .((.((.((.((((((((......)))))).)).)).)).))((((((......((((((.((.((.((.(((((((....).)))))).....)).)).))))))))......)))))) ( -45.30) >DroWil_CAF1 64136 111 + 1 CGAAGGUCCUUGGUGGAAUAACUCCACGGCCUCCCCUCCCCAAUGG---AACACCACAGCAUUACCUCCCCAAUUG---AACACCACAACCGAUCGUCCUUGGUGG---AACAACUCGAC ((((((((....(((((.....))))))))))....(((((((.((---(..........................---.................))))))).))---).....))).. ( -20.43) >DroYak_CAF1 87728 120 + 1 UCAAGGUCCAUGGUGGGGAAACACCACCACUCAAGGUCCAUGGUGGGGAAACAACUCAACGGAAGGUCCAUGGUGGGGAAAUAACUCUACUCAAGGUCCAUGGUGGGGAAACAACUCAAC ......(((.(((((((....).))))))........((((.(((((....).......(....)..((.((((((((......)))))).)).)).)))).)))))))........... ( -39.70) >consensus UCAAGGUCCAUGGUGGGGAAACACCACUACUCAAGGUCCAUGGUGGGGAAACACCACAACUCAAGGUCCAUGGUGGGGAAACACCACUACUCAAGGUCCAUGGUGGGGAAACAACACUAC ......(((((.(((((.........(((((...((.((.........................)).))..)))))(....)..............))))).)))))............. (-13.72 = -14.81 + 1.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:19 2006