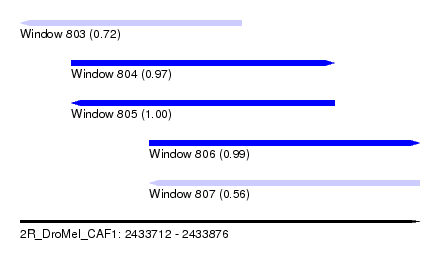
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,433,712 – 2,433,876 |
| Length | 164 |
| Max. P | 0.997974 |

| Location | 2,433,712 – 2,433,803 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 88.91 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -19.08 |
| Energy contribution | -19.64 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2433712 91 - 20766785 AGCUGGAAUCCAUUACUGCAAUCACUGGGUGCAUUGUUUGC-UGCAUUUUUCAGCUUGCACACAGUGCCACAC----ACACGUGUGUCUAGAUGU------A ..(((((...............(((((.(((((......((-((.......)))).))))).))))).(((((----....))))))))))....------. ( -26.80) >DroSec_CAF1 89614 94 - 1 AGCUGGAAUCCAUUACUGCAAUCACUGGGUGCAUUGUUUGCAUGCAUUUUUCAGCUCGCACACAGUGCCACACA--CACACGUGUGUCUAGAUGU------A ..((((((((((.............)))))(((((((.(((..((........))..))).)))))))..((((--(....))))))))))....------. ( -27.22) >DroSim_CAF1 47103 96 - 1 AGCUGGAAUCCAUUACUGCAAUCACUGGGUGCAUUGUUUGCAUGCAUUUUUCAGCUCGCACACAGUGCCACACACGCACACGUGUGUUUAGAUGU------A ..((((((((((.............)))))(((((((.(((..((........))..))).)))))))..((((((....)))))))))))....------. ( -26.52) >DroYak_CAF1 45933 98 - 1 AGUUCGAAUCCAUUACUGCAAGCACUGUGUGCAUUGUUUGCAUUCAUUUUUCAGCUCGCACACAGUGCAAAAC----ACACGUGCGUCUAGAUGUACGAGUA .............((((....(((((((((((...(((..............)))..))))))))))).....----...(((((((....))))))))))) ( -29.94) >consensus AGCUGGAAUCCAUUACUGCAAUCACUGGGUGCAUUGUUUGCAUGCAUUUUUCAGCUCGCACACAGUGCCACAC____ACACGUGUGUCUAGAUGU______A ((((((((........(((((.((.((....)).)).)))))......)))))))).((((((.(((...........)))))))))............... (-19.08 = -19.64 + 0.56)

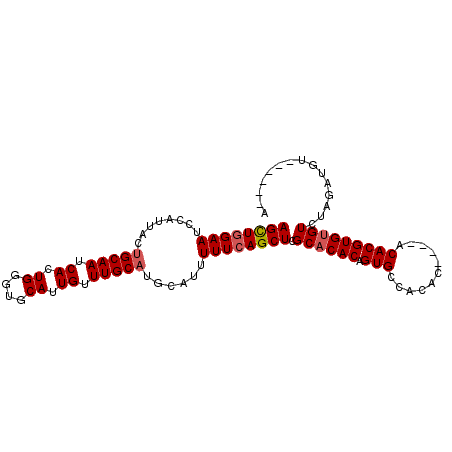
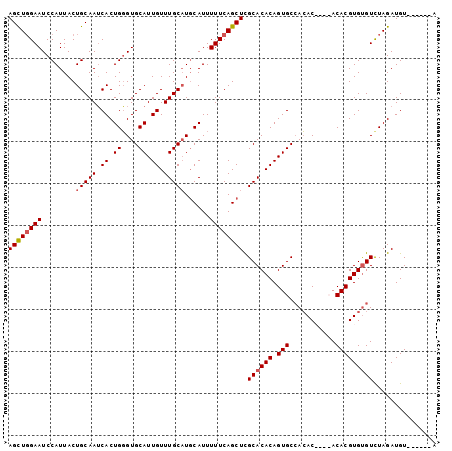
| Location | 2,433,733 – 2,433,841 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.97 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -25.30 |
| Energy contribution | -26.46 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2433733 108 + 20766785 GUGGCACUGUGUGCAAGCUGAAAAAUGCA-GCAAACAAUGCACCCAGUGAUUGCAGUAAUGGAUUCCAGCUACUGUAAACACCUGGG--AAAUGUCAGAGGCGUCUGAAUG ...(((.(((.(((..((........)).-))).))).))).(((((((.((((((((.(((...)))..)))))))).)).)))))--.....(((((....)))))... ( -36.60) >DroSec_CAF1 89637 109 + 1 GUGGCACUGUGUGCGAGCUGAAAAAUGCAUGCAAACAAUGCACCCAGUGAUUGCAGUAAUGGAUUCCAGCUACUGCAAACACCUGGG--CAGGGUCAGAGGCGUCUGAAUG ...(((.(((.((((.((........)).)))).))).))).(((((((.((((((((.(((...)))..)))))))).)).)))))--.....(((((....)))))... ( -40.60) >DroSim_CAF1 47128 109 + 1 GUGGCACUGUGUGCGAGCUGAAAAAUGCAUGCAAACAAUGCACCCAGUGAUUGCAGUAAUGGAUUCCAGCUACUGCAAACACCUGGG--CAGGGUCAGAGGCGUCUGAAUG ...(((.(((.((((.((........)).)))).))).))).(((((((.((((((((.(((...)))..)))))))).)).)))))--.....(((((....)))))... ( -40.60) >DroEre_CAF1 45343 100 + 1 -GUGCAC-GUGUGCGAGCUGAAAAAUGGAUGCAAACAAUGCACGCAGUGAUUGCAGUAAUGUA-------CACUGCAAACAACUGGG--AAGGGUCAGAGGCGACCGAGUG -(((((.-((.((((..(........)..)))).))..)))))((((((..((((....))))-------))))))...........--...((((......))))..... ( -29.30) >DroYak_CAF1 45960 102 + 1 UUUGCACUGUGUGCGAGCUGAAAAAUGAAUGCAAACAAUGCACACAGUGCUUGCAGUAAUGGAUUCGAACUACUGUAAACAACUGGGGAAAGGGUCAGAUAC--------- ...((((((((((((.((............))......))))))))))))((((((((.(........).))))))))....((((........))))....--------- ( -28.40) >consensus GUGGCACUGUGUGCGAGCUGAAAAAUGCAUGCAAACAAUGCACCCAGUGAUUGCAGUAAUGGAUUCCAGCUACUGCAAACACCUGGG__AAGGGUCAGAGGCGUCUGAAUG ...(((.(((.((((.((........)).)))).))).))).(((((((.((((((((............)))))))).)).)))))........................ (-25.30 = -26.46 + 1.16)

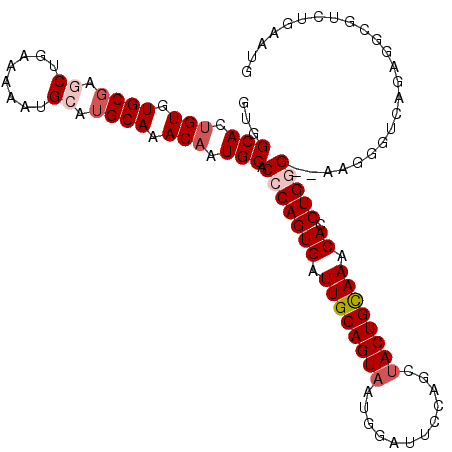
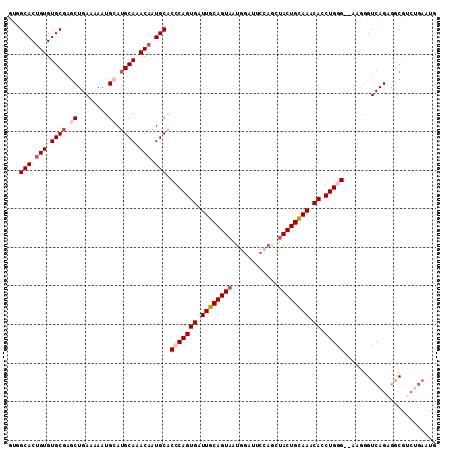
| Location | 2,433,733 – 2,433,841 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.97 |
| Mean single sequence MFE | -35.35 |
| Consensus MFE | -25.76 |
| Energy contribution | -28.48 |
| Covariance contribution | 2.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.78 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
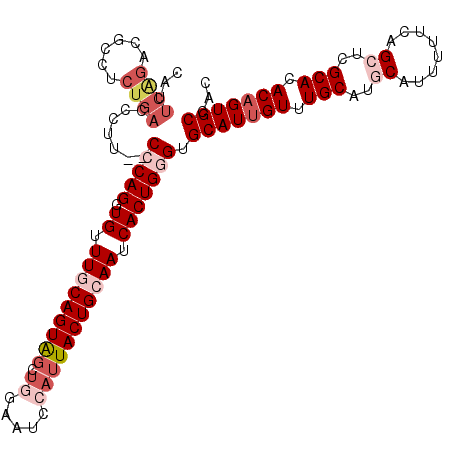
>2R_DroMel_CAF1 2433733 108 - 20766785 CAUUCAGACGCCUCUGACAUUU--CCCAGGUGUUUACAGUAGCUGGAAUCCAUUACUGCAAUCACUGGGUGCAUUGUUUGC-UGCAUUUUUCAGCUUGCACACAGUGCCAC ...(((((....))))).....--(((((.((.((.((((((.(((...))))))))).)).))))))).(((((((.(((-.((........))..))).)))))))... ( -34.70) >DroSec_CAF1 89637 109 - 1 CAUUCAGACGCCUCUGACCCUG--CCCAGGUGUUUGCAGUAGCUGGAAUCCAUUACUGCAAUCACUGGGUGCAUUGUUUGCAUGCAUUUUUCAGCUCGCACACAGUGCCAC ...(((((....)))))....(--(((((.((.(((((((((.(((...)))))))))))).))))))))(((((((.(((..((........))..))).)))))))... ( -43.90) >DroSim_CAF1 47128 109 - 1 CAUUCAGACGCCUCUGACCCUG--CCCAGGUGUUUGCAGUAGCUGGAAUCCAUUACUGCAAUCACUGGGUGCAUUGUUUGCAUGCAUUUUUCAGCUCGCACACAGUGCCAC ...(((((....)))))....(--(((((.((.(((((((((.(((...)))))))))))).))))))))(((((((.(((..((........))..))).)))))))... ( -43.90) >DroEre_CAF1 45343 100 - 1 CACUCGGUCGCCUCUGACCCUU--CCCAGUUGUUUGCAGUG-------UACAUUACUGCAAUCACUGCGUGCAUUGUUUGCAUCCAUUUUUCAGCUCGCACAC-GUGCAC- .....(((((....)))))...--..((((.(.((((((((-------.....)))))))).))))).((((((.((.(((................))).))-))))))- ( -28.29) >DroYak_CAF1 45960 102 - 1 ---------GUAUCUGACCCUUUCCCCAGUUGUUUACAGUAGUUCGAAUCCAUUACUGCAAGCACUGUGUGCAUUGUUUGCAUUCAUUUUUCAGCUCGCACACAGUGCAAA ---------(((..((((..........))))..))).(((((...........)))))..(((((((((((...(((..............)))..)))))))))))... ( -25.94) >consensus CAUUCAGACGCCUCUGACCCUU__CCCAGGUGUUUGCAGUAGCUGGAAUCCAUUACUGCAAUCACUGGGUGCAUUGUUUGCAUGCAUUUUUCAGCUCGCACACAGUGCCAC ...((((......)))).......(((((.((.(((((((((.((.....))))))))))).))))))).(((((((.(((..((........))..))).)))))))... (-25.76 = -28.48 + 2.72)

| Location | 2,433,765 – 2,433,876 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.98 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -22.04 |
| Energy contribution | -23.40 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
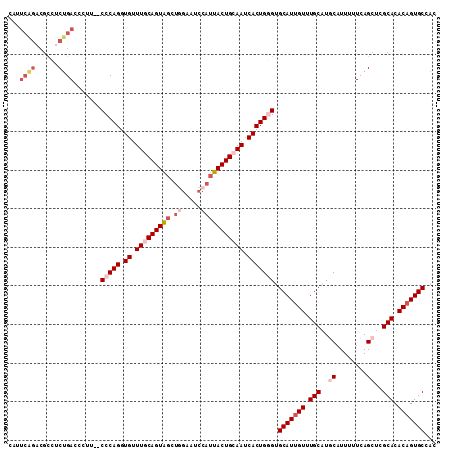
>2R_DroMel_CAF1 2433765 111 + 20766785 AACAAUGCACCCAGUGAUUGCAGUAAUGGAUUCCAGCUACUGUAAACACCUGGG--AAAUGUCAGAGGCGUCUGAAUGGCCC--CAAAAUAGUAUUGAGUAUUGUAUUCGUCCAU .(((((((.(((((((.((((((((.(((...)))..)))))))).)).)))))--..........((.(((.....))).)--).............))))))).......... ( -34.20) >DroSec_CAF1 89670 111 + 1 AACAAUGCACCCAGUGAUUGCAGUAAUGGAUUCCAGCUACUGCAAACACCUGGG--CAGGGUCAGAGGCGUCUGAAUGCCCC--CAAAAUCGAAUUGAGUAUUGUAUUCGUCCAA .(((((((.(((((((.((((((((.(((...)))..)))))))).)).)))))--..(((.....(((((....)))))))--).............))))))).......... ( -39.00) >DroSim_CAF1 47161 111 + 1 AACAAUGCACCCAGUGAUUGCAGUAAUGGAUUCCAGCUACUGCAAACACCUGGG--CAGGGUCAGAGGCGUCUGAAUGCCCC--CAAAAUAGUAUUGAGUAUUGUAUUCGUCCAU .(((((((.(((((((.((((((((.(((...)))..)))))))).)).)))))--..(((.....(((((....)))))))--).............))))))).......... ( -39.00) >DroEre_CAF1 45374 99 + 1 AACAAUGCACGCAGUGAUUGCAGUAAUGUA-------CACUGCAAACAACUGGG--AAGGGUCAGAGGCGACCGAGUGGCCCCCGAAAAUA-------GUAUUGUAUUCGUCCAU .(((((((..((((((..((((....))))-------)))))).........((--..((((((..((...))...)))))))).......-------))))))).......... ( -31.40) >DroYak_CAF1 45993 93 + 1 AACAAUGCACACAGUGCUUGCAGUAAUGGAUUCGAACUACUGUAAACAACUGGGGAAAGGGUCAGAUAC---------------CAAAAUA-------GUAUUGUAUUCGUCCAU .(((((((...((((..((((((((.(........).))))))))...))))......((........)---------------)......-------))))))).......... ( -21.40) >consensus AACAAUGCACCCAGUGAUUGCAGUAAUGGAUUCCAGCUACUGCAAACACCUGGG__AAGGGUCAGAGGCGUCUGAAUGCCCC__CAAAAUAG_AUUGAGUAUUGUAUUCGUCCAU .(((((((.(((((((.((((((((............)))))))).)).))))).......((((......)))).......................))))))).......... (-22.04 = -23.40 + 1.36)

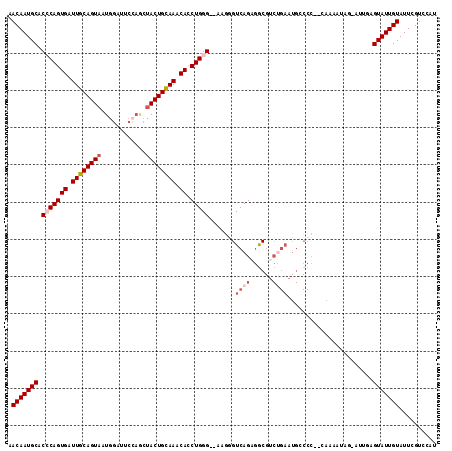
| Location | 2,433,765 – 2,433,876 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.98 |
| Mean single sequence MFE | -30.44 |
| Consensus MFE | -16.66 |
| Energy contribution | -18.78 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
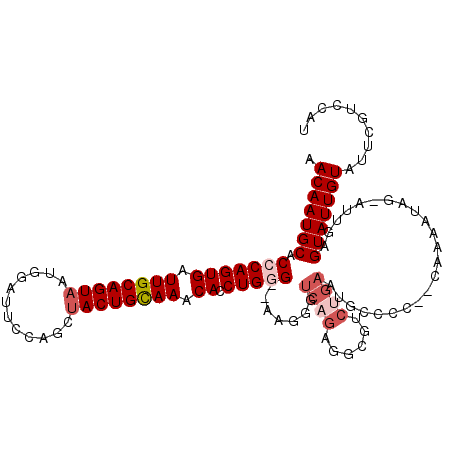
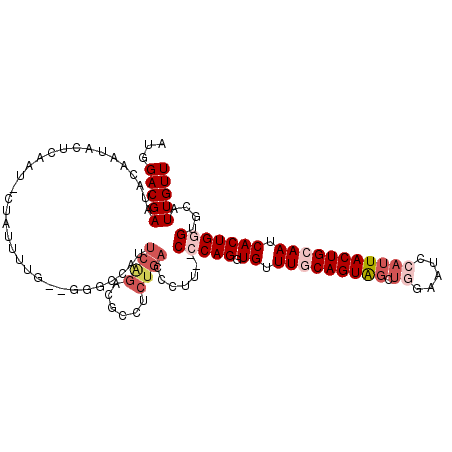
>2R_DroMel_CAF1 2433765 111 - 20766785 AUGGACGAAUACAAUACUCAAUACUAUUUUG--GGGCCAUUCAGACGCCUCUGACAUUU--CCCAGGUGUUUACAGUAGCUGGAAUCCAUUACUGCAAUCACUGGGUGCAUUGUU ...(((((........(((((.......)))--))((...(((((....))))).....--(((((.((.((.((((((.(((...))))))))).)).))))))).)).))))) ( -27.20) >DroSec_CAF1 89670 111 - 1 UUGGACGAAUACAAUACUCAAUUCGAUUUUG--GGGGCAUUCAGACGCCUCUGACCCUG--CCCAGGUGUUUGCAGUAGCUGGAAUCCAUUACUGCAAUCACUGGGUGCAUUGUU ..((.(((((..........)))))...(..--(((((........)))))..).)).(--(((((.((.(((((((((.(((...)))))))))))).))))))))........ ( -38.80) >DroSim_CAF1 47161 111 - 1 AUGGACGAAUACAAUACUCAAUACUAUUUUG--GGGGCAUUCAGACGCCUCUGACCCUG--CCCAGGUGUUUGCAGUAGCUGGAAUCCAUUACUGCAAUCACUGGGUGCAUUGUU ...(((((....................(..--(((((........)))))..)....(--(((((.((.(((((((((.(((...)))))))))))).))))))))...))))) ( -37.20) >DroEre_CAF1 45374 99 - 1 AUGGACGAAUACAAUAC-------UAUUUUCGGGGGCCACUCGGUCGCCUCUGACCCUU--CCCAGUUGUUUGCAGUG-------UACAUUACUGCAAUCACUGCGUGCAUUGUU (((.(((..........-------.....((((((((.((...)).)))))))).....--..((((.(.((((((((-------.....)))))))).)))))))).))).... ( -30.60) >DroYak_CAF1 45993 93 - 1 AUGGACGAAUACAAUAC-------UAUUUUG---------------GUAUCUGACCCUUUCCCCAGUUGUUUACAGUAGUUCGAAUCCAUUACUGCAAGCACUGUGUGCAUUGUU (((.(((....((((((-------(.....)---------------)))).))..........((((.((((.(((((((........))))))).))))))))))).))).... ( -18.40) >consensus AUGGACGAAUACAAUACUCAAU_CUAUUUUG__GGGCCAUUCAGACGCCUCUGACCCUU__CCCAGGUGUUUGCAGUAGCUGGAAUCCAUUACUGCAAUCACUGGGUGCAUUGUU ...(((((................................((((......)))).......(((((.((.(((((((((.((.....))))))))))).)))))))....))))) (-16.66 = -18.78 + 2.12)

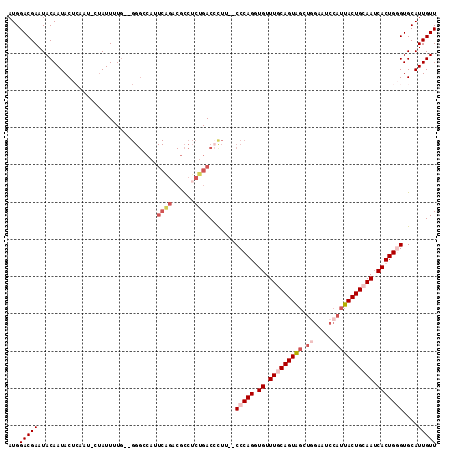
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:59 2006