
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,526,234 – 13,526,336 |
| Length | 102 |
| Max. P | 0.695826 |

| Location | 13,526,234 – 13,526,336 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -22.20 |
| Consensus MFE | -12.49 |
| Energy contribution | -13.77 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13526234 102 - 20766785 GGGCUAAAAGGUAGAUGCGA-AAAAUACCAG--CACGUUAAAUUGAAUUUAACGUGAAAUGCAGUGAGAUGAAAUGAAAUUUUCGCUGUCGCACUGUCUAACAAG- ........((..((.(((((-.......((.--((((((((((...))))))))))...))((((((((...........)))))))))))))))..))......- ( -28.60) >DroVir_CAF1 48995 93 - 1 GGGCUA------AAAAAUGA-AAAAUACCAG--CACGUUAAAUUGAAUUUAACGUGAAAUGCAAUGAAACGUAAUGAAAUUUUCGCUGUCAG-UCGUC--UCAAG- .((((.------.....(((-(((.......--((((((((((...))))))))))...(((........)))......)))))).....))-))...--.....- ( -14.90) >DroGri_CAF1 42832 92 - 1 AGG--A------AAAAGUGAGAAAAUACCAG--CACGUUGAAUUGAAUUUAACGUGAAAUGCAAUGAAAUGUAAUGAAAUUUUCGCCGUCAG-UAGUC--UCAAG- ...--.------.....(((((...(((...--((((((((((...))))))))))...((((......))))..................)-)).))--)))..- ( -17.80) >DroEre_CAF1 75026 102 - 1 GGGGUAAAAGGUGGACGGGC-GAAAUACCAG--CACGUUAAAUUGAAUUUCACGUAAAAUGCAGUGAAAUGAAAUGAAAUUUUCGCUGUCGCACUGACUAACAAG- ..(((....(((((((((.(-((((......--...(((..(((..(((((((((.....)).)))))))..)))..))))))))))))).)))).)))......- ( -24.30) >DroAna_CAF1 75385 103 - 1 GGUGAAUAAGGUAGGAGUGA-AAAAUACCAG--CACGUUAAAUUGAAUUUAACGUGAAAUGCAGUGAAACGAAAUGAAAUUUUCGCUGUCGCAAUGUCUAACAAGG .((((....((((....(..-..).))))..--((((((((((...)))))))))).....((((((((...........)))))))))))).............. ( -22.80) >DroPer_CAF1 80044 101 - 1 AG---ACUAGGUGGAAGUGA-GAAGUACAAGCGCACGUUAAAUUGAAUUUAACGUGAAAUUCAGCCAAACGAAAUGAAAUUUUUCCUGUCGCAGUGUCUAACAAG- ((---((...(((..((.((-(((((.......((((((((((...))))))))))...((((...........)))))))))))))..)))...))))......- ( -24.80) >consensus GGG_UA_AAGGUAGAAGUGA_AAAAUACCAG__CACGUUAAAUUGAAUUUAACGUGAAAUGCAGUGAAACGAAAUGAAAUUUUCGCUGUCGCACUGUCUAACAAG_ ................(((..............((((((((((...))))))))))....(((((((((...........)))))))))))).............. (-12.49 = -13.77 + 1.28)

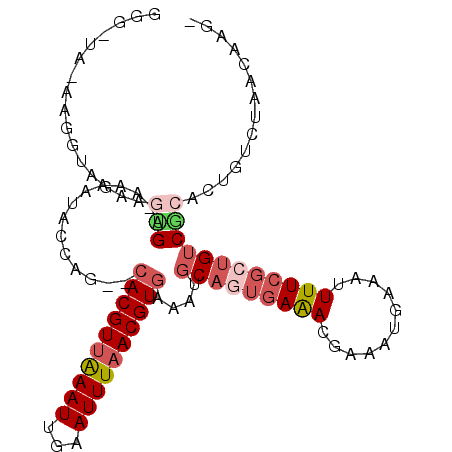
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:10 2006