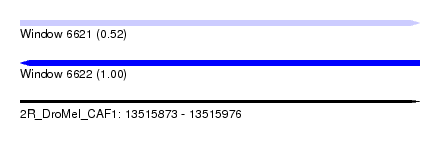
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,515,873 – 13,515,976 |
| Length | 103 |
| Max. P | 0.999077 |

| Location | 13,515,873 – 13,515,976 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -17.85 |
| Energy contribution | -20.98 |
| Covariance contribution | 3.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
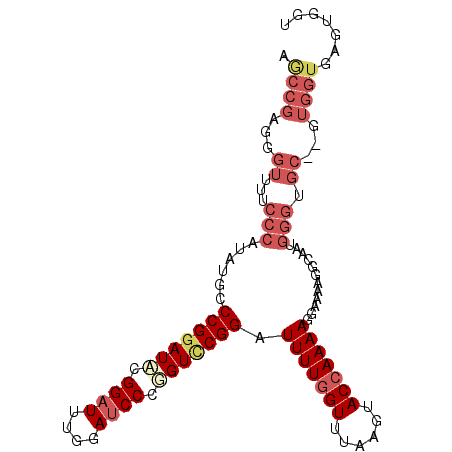
>2R_DroMel_CAF1 13515873 103 + 20766785 AACCGACGGUUUUCCCAUAUGCCCGGAUACGGAUUUGGAUCCUGGUCCGGAUUUUGGUUUAAGUACGAAAAGGAAAAGGCGAUGGGUGCGAGUGGUGAGUGGU .((((...((...(((((.(((((((((..((((....))))..)))))..((((.((......)).))))......))))))))).))...))))....... ( -30.80) >DroSec_CAF1 64331 101 + 1 AGCCGAGGGUUUUCCCAUAUGCCCGGAUCCGGAUUUGGAUCCCAGUUCGGAUUUUGGUUUAAGUACCAAAAGGAAAAGGCGAUGGGUGC--GUGGUGAGUGGU .((((...((...(((((.((((.(((((((....)))))))...(((...(((((((......))))))).)))..))))))))).))--.))))....... ( -34.30) >DroSim_CAF1 65059 101 + 1 AGCCGAGGGUUUUCCCAUAUGCCCGGAUCCGGAUUUGGAUCCCGGUUCGGAUUUUGGUUUAAGUACCAAAAGGAAAAGGAAAAGGGUGC--GUGGUGAGUGGU .((((......(((((((..(((((((((((....)))))))(..(((...(((((((......))))))).)))..).....))))..--)))).))))))) ( -34.00) >DroYak_CAF1 67195 81 + 1 AGCCGGAGGAUUUCCCAUAUGCCCGUAUACGGAUUGGGAACCGUGUCCGGAUUUUGGUUUAAGUACCAAAAUGCUAAAGGA---------------------- ..(((((.(..((((((.....(((....)))..))))))...).)))))((((((((......)))))))).........---------------------- ( -28.20) >consensus AGCCGAGGGUUUUCCCAUAUGCCCGGAUACGGAUUUGGAUCCCGGUCCGGAUUUUGGUUUAAGUACCAAAAGGAAAAGGCAAUGGGUGC__GUGGUGAGUGGU .((((...((...(((......(((((((.((((....)))).))))))).(((((((......)))))))............))).))...))))....... (-17.85 = -20.98 + 3.12)

| Location | 13,515,873 – 13,515,976 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -25.85 |
| Consensus MFE | -16.76 |
| Energy contribution | -17.82 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.65 |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.999077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
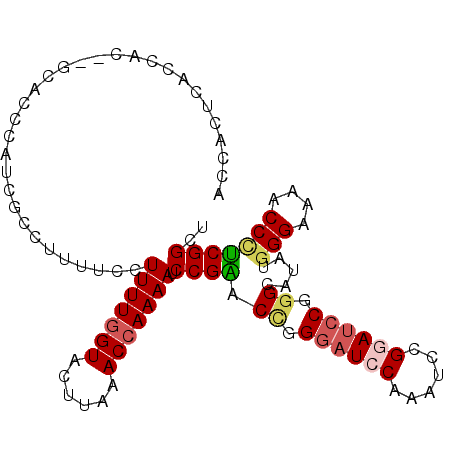
>2R_DroMel_CAF1 13515873 103 - 20766785 ACCACUCACCACUCGCACCCAUCGCCUUUUCCUUUUCGUACUUAAACCAAAAUCCGGACCAGGAUCCAAAUCCGUAUCCGGGCAUAUGGGAAAACCGUCGGUU ..................((..((..((((((((((.((......)).))))((((((...((((....))))...)))))).....))))))..))..)).. ( -21.50) >DroSec_CAF1 64331 101 - 1 ACCACUCACCAC--GCACCCAUCGCCUUUUCCUUUUGGUACUUAAACCAAAAUCCGAACUGGGAUCCAAAUCCGGAUCCGGGCAUAUGGGAAAACCCUCGGCU ............--...(((((.((((.....(((((((......))))))).........((((((......))))))))))..)))))............. ( -28.40) >DroSim_CAF1 65059 101 - 1 ACCACUCACCAC--GCACCCUUUUCCUUUUCCUUUUGGUACUUAAACCAAAAUCCGAACCGGGAUCCAAAUCCGGAUCCGGGCAUAUGGGAAAACCCUCGGCU ............--..................(((((((......))))))).((((.((.((((((......)))))).)).....(((....))))))).. ( -27.50) >DroYak_CAF1 67195 81 - 1 ----------------------UCCUUUAGCAUUUUGGUACUUAAACCAAAAUCCGGACACGGUUCCCAAUCCGUAUACGGGCAUAUGGGAAAUCCUCCGGCU ----------------------......(((((((((((......)))))))).((((.....((((((.((((....))))....))))))....))))))) ( -26.00) >consensus ACCACUCACCAC__GCACCCAUCGCCUUUUCCUUUUGGUACUUAAACCAAAAUCCGAACCGGGAUCCAAAUCCGGAUCCGGGCAUAUGGGAAAACCCUCGGCU ................................(((((((......))))))).((((.((.((((((......)))))).)).....(((....))))))).. (-16.76 = -17.82 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:05 2006