| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,500,886 – 13,500,995 |
| Length | 109 |
| Max. P | 0.749850 |

| Location | 13,500,886 – 13,500,995 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 83.02 |
| Mean single sequence MFE | -19.64 |
| Consensus MFE | -14.11 |
| Energy contribution | -14.27 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
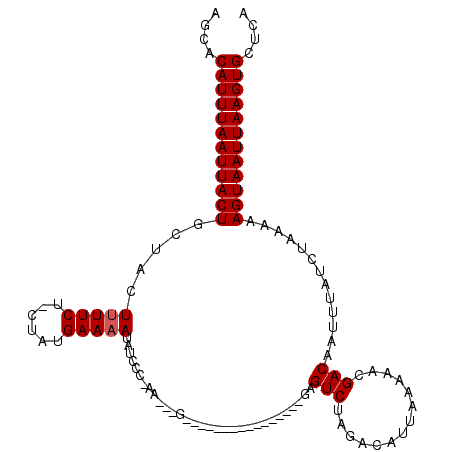
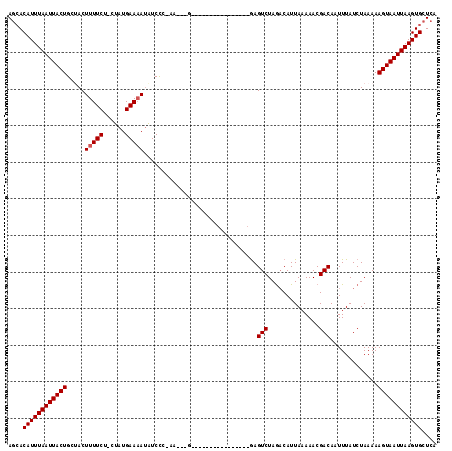
>2R_DroMel_CAF1 13500886 109 + 20766785 AGCACAUUUAAUUACUGCUACUUUUCU-CUAUGAA-AGAUCCC-AGU--CAG---GGACAGGAGGCGAGUCUAGACAUUAAAAACGACAAUUUAUCUAAAAAGUAAUUAAGUGCUCA (((((..((((((((((((.(((.(((-((.((..-......)-)..--.))---))).))).)))..(((..............))).............)))))))))))))).. ( -23.94) >DroPse_CAF1 46953 113 + 1 AGCACAUUUAAUUACUGCUACUUUUCU-CUAUGAAAAUGUCCC-AAAGCGAGCAA--GCGAGAGGCGAGUCUAGACAUUAAAAACGACAAUUUAUCUAAAAAGUAAUUAAGUGCUCA (((((..((((((((((((.....(((-((((....)))....-...((......--)))))))))..(((..............))).............)))))))))))))).. ( -21.44) >DroGri_CAF1 12283 94 + 1 AUCACAUUUAAUUACUGCCACUUUUCU-CUAUGAAAAUAUCUA-A---------------------GAGUCUAGCCAUUAAAAACGACAAUUUAUCUAAAAAGUAAUUAAGUGCUUA ....((((((((((((.....(((((.-....)))))......-.---------------------..(((..............))).............)))))))))))).... ( -14.34) >DroWil_CAF1 16039 99 + 1 AGAACAUUUAAUUACUGCUACUUUUCU-CUAUGAAAAUAUCUC-UAAGAG----------------AAGUCUAGACAUUAAAAACGACAAUUUAUCUAAAAAGUAAUUAAGUGCUCA .((.((((((((((((.(((..(((((-((..((.......))-..))))----------------)))..)))....((((........)))).......)))))))))))).)). ( -20.60) >DroMoj_CAF1 16190 96 + 1 AGCACAUUUAAUUACUGCCACUUUUCUACUAUGAAAAUAUCUAAA---------------------GAGUCUAAACAUUAAAAACGACAAUUUAUCUAAAAAGUAAUUAAGUGCUCA (((((..(((((((((.....(((((......)))))....((((---------------------..(((..............)))..)))).......)))))))))))))).. ( -16.24) >DroPer_CAF1 55022 113 + 1 AGCACAUUUAAUUACUGCUACUUUUCU-CUAUGAAAAUGUCCC-AAUGCGAGCAA--GCGAGAGGCGAGUCUAGACAUUAAAAACGACAAUUUAUCUAAAAAGUAAUUAAGUGCUCA (((((..(((((((((...((((.(((-((((....)).....-..(((......--)))))))).)))).((((...((((........))))))))...)))))))))))))).. ( -21.30) >consensus AGCACAUUUAAUUACUGCUACUUUUCU_CUAUGAAAAUAUCCC_AA___G________________GAGUCUAGACAUUAAAAACGACAAUUUAUCUAAAAAGUAAUUAAGUGCUCA ....((((((((((((.....(((((......)))))...............................(((..............))).............)))))))))))).... (-14.11 = -14.27 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:56 2006