| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
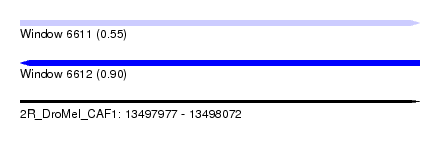
| Location | 13,497,977 – 13,498,072 |
| Length | 95 |
| Max. P | 0.903309 |

| Location | 13,497,977 – 13,498,072 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 90.25 |
| Mean single sequence MFE | -20.67 |
| Consensus MFE | -15.60 |
| Energy contribution | -16.43 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13497977 95 + 20766785 AAAUGGAGCUCAAUUCAGCGGAAAUGCAUAUUAAGUGCAAUUACUGCAUAAUUAACCAGUGAUUCUGCAAUCACUAAUUGCAACCCACACCAAAA ...(((.(((......)))((...((((......(((((.....)))))........(((((((....)))))))...))))..))...)))... ( -20.90) >DroSec_CAF1 46486 95 + 1 AAAUCAAGCUCAAUUCAGCGGAAAUGCAUAUUAAGUGCAAUUACUGCAUAAUUAACCAGUGAUUCUGCAAUCACUAAUUGCAACUCACACCGAAU ............((((.((((((.(((((.....))))).)).))))...........((((...((((((.....))))))..))))...)))) ( -21.20) >DroSim_CAF1 46773 95 + 1 AAAUGGAGCUCGAUUCAGCGGAAAUGCAUAUUAAGUGCAAUUACUGCAUAAUUAACCAGUGAUUCUGCAAUCACUAAUUGCAACUCACACCGAAU ...(((...........((((((.(((((.....))))).)).))))...........((((...((((((.....))))))..)))).)))... ( -21.90) >DroEre_CAF1 47672 95 + 1 AAAUUGAGCUCAAUUCAGCGGAAAUGCAUAUUAAGUGCUAUUACUGCAUAAUUAACCAGUGAUUCUGCAAUCACUAAUUGCAACUCACACUGAAU .(((((....)))))..((((.(((((((.....)))).))).)))).........(((((....((((((.....)))))).....)))))... ( -21.70) >DroYak_CAF1 48906 95 + 1 AAAUUGAGCUCAAUUCAGCGGAAAUGCAUAUUAAGUGCAAUUACUGCAUAAUUAACCAGUGAUUCUGCAAUCACUAAUUGCAACCCACACUGAGU .(((((....)))))..((((((.(((((.....))))).)).)))).......(((((((....((((((.....)))))).....))))).)) ( -22.50) >DroAna_CAF1 48306 89 + 1 UAAUAGAGCUCAAUUCAGCGGAAAUGCAUAUUAAGUGCAAUUACUGCAUAAUUAACCAGGCACUUAAAAAACACUAAUUGCAACUCUCU------ ....((((.........((((((.(((((.....))))).)).))))............(((.(((........))).)))..))))..------ ( -15.80) >consensus AAAUGGAGCUCAAUUCAGCGGAAAUGCAUAUUAAGUGCAAUUACUGCAUAAUUAACCAGUGAUUCUGCAAUCACUAAUUGCAACUCACACCGAAU .......((........((((((.(((((.....))))).)).))))..........(((((((....)))))))....)).............. (-15.60 = -16.43 + 0.83)

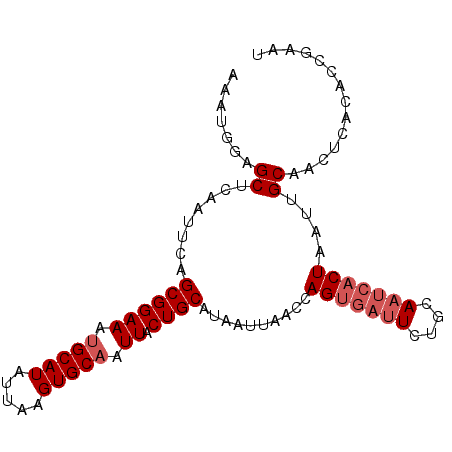

| Location | 13,497,977 – 13,498,072 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 90.25 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -21.12 |
| Energy contribution | -22.32 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13497977 95 - 20766785 UUUUGGUGUGGGUUGCAAUUAGUGAUUGCAGAAUCACUGGUUAAUUAUGCAGUAAUUGCACUUAAUAUGCAUUUCCGCUGAAUUGAGCUCCAUUU .((..(((.(((.((((((((((((((....))))))))))).....(((((...)))))........))).))))))..))............. ( -23.50) >DroSec_CAF1 46486 95 - 1 AUUCGGUGUGAGUUGCAAUUAGUGAUUGCAGAAUCACUGGUUAAUUAUGCAGUAAUUGCACUUAAUAUGCAUUUCCGCUGAAUUGAGCUUGAUUU ((((((((.(((.((((((((((((((....))))))))))).....(((((...)))))........))).)))))))))))............ ( -27.30) >DroSim_CAF1 46773 95 - 1 AUUCGGUGUGAGUUGCAAUUAGUGAUUGCAGAAUCACUGGUUAAUUAUGCAGUAAUUGCACUUAAUAUGCAUUUCCGCUGAAUCGAGCUCCAUUU ((((((((.(((.((((((((((((((....))))))))))).....(((((...)))))........))).)))))))))))............ ( -27.30) >DroEre_CAF1 47672 95 - 1 AUUCAGUGUGAGUUGCAAUUAGUGAUUGCAGAAUCACUGGUUAAUUAUGCAGUAAUAGCACUUAAUAUGCAUUUCCGCUGAAUUGAGCUCAAUUU ((((((((.(((.((((((((((((((....))))))))))).....(((.......)))........))).)))))))))))............ ( -27.10) >DroYak_CAF1 48906 95 - 1 ACUCAGUGUGGGUUGCAAUUAGUGAUUGCAGAAUCACUGGUUAAUUAUGCAGUAAUUGCACUUAAUAUGCAUUUCCGCUGAAUUGAGCUCAAUUU .(((((((((((.((((((((((((((....))))))))))).....(((((...)))))........)))..)))))...))))))........ ( -26.90) >DroAna_CAF1 48306 89 - 1 ------AGAGAGUUGCAAUUAGUGUUUUUUAAGUGCCUGGUUAAUUAUGCAGUAAUUGCACUUAAUAUGCAUUUCCGCUGAAUUGAGCUCUAUUA ------..((((((.(((((((((......((((((...(((((...(((((...))))).)))))..)))))).))))).)))))))))).... ( -21.00) >consensus AUUCGGUGUGAGUUGCAAUUAGUGAUUGCAGAAUCACUGGUUAAUUAUGCAGUAAUUGCACUUAAUAUGCAUUUCCGCUGAAUUGAGCUCAAUUU .(((((((.(((.((((((((((((((....))))))))))).....(((((...)))))........))).))))))))))............. (-21.12 = -22.32 + 1.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:55 2006