
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,495,232 – 13,495,337 |
| Length | 105 |
| Max. P | 0.754451 |

| Location | 13,495,232 – 13,495,337 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.84 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -13.45 |
| Energy contribution | -12.48 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
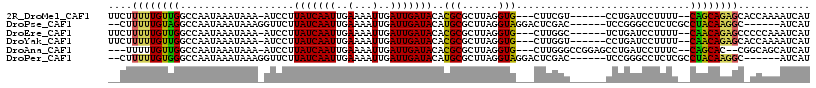
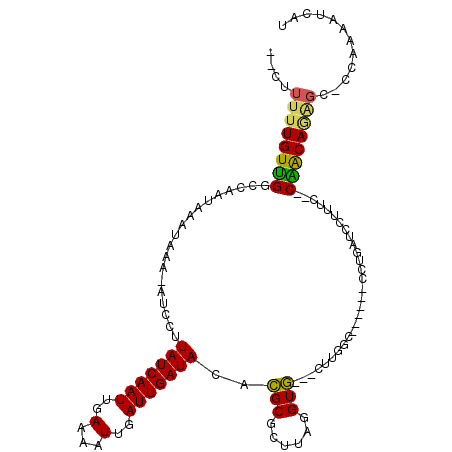
>2R_DroMel_CAF1 13495232 105 - 20766785 UUCUUUUUGUUGGCCAAUAAAUAAA-AUCCUUAUCAAUUGAAAAUUGAUUGAUACACGCGCUUAGGUG---CUUCGU------CCUGAUCCUUUU--CAGCAGAGCACCAAAAUCAU ....(((((((........))))))-)....(((((((..(...)..)))))))..........((((---(((...------.((((......)--)))..)))))))........ ( -22.80) >DroPse_CAF1 41053 103 - 1 --CUUUUUGUAGGCCAAUAAAUAAAGGUUCUUAUCAAUUGAAAAUUGAUUGAUACAUGCGCUUAGGUAGGACUCGAC------UCCGGGCCUCUCGCCUACAAGGC------AUCAU --..((((((((((.........(((((...(((((((..(...)..)))))))...)).))).(..(((.((((..------..)))))))..))))))))))).------..... ( -30.20) >DroEre_CAF1 44873 105 - 1 UUCUUUUUGUUGGCCAAUAAAUAAA-AUCCUUAUCAAUUGAAAAUUGAUUGAUACACGCGCUUAGGUG---CUUGGC------UCUGAUCCUUUU--CAACAGAGCCCCCAAAUCAU .........((((............-.....(((((((..(...)..)))))))...((((....)))---)..(((------((((........--...))))))).))))..... ( -24.00) >DroYak_CAF1 45930 105 - 1 UUCUUUUUGUUGGCCAAUAAAUAAA-AUCCUUAUCAAUUGAAAAUUGAUUGAUACACGCGCUUAGGUG---CUUGGU------CCUGAUCCUUUU--CAACAGAGCACCAAAAUCAU ....(((((((........))))))-)....(((((((..(...)..)))))))..........((((---(((.((------..(((......)--)))).)))))))........ ( -21.70) >DroAna_CAF1 45473 106 - 1 ---UUUUUGUUGGCCAAUAAAUAAA-AUCCUUAUCAAUUGAAAAUUGAUUGAUACACGCGCUUAGGUG---CUUGGGCCGGAGCCUGAUCCUUUC--CAGCAC--CGGCAGCAUCAU ---....(((((.((..........-.....(((((((..(...)..)))))))..........((((---((.(((..(((......)))..))--))))))--)))))))).... ( -31.40) >DroPer_CAF1 49119 103 - 1 --CUUUUUGUGGGCCAAUAAAUAAAGGUUCUUAUCAAUUGAAAAUUGAUUGAUACAUGCGCUUAGGUAGGACUCGAC------UCCGGGCCUCUCGCCUACAAGGC------AUCAU --..((((((((((.........(((((...(((((((..(...)..)))))))...)).))).(..(((.((((..------..)))))))..))))))))))).------..... ( -29.90) >consensus __CUUUUUGUUGGCCAAUAAAUAAA_AUCCUUAUCAAUUGAAAAUUGAUUGAUACACGCGCUUAGGUG___CUUGGC______CCUGAUCCUUUC__CAACAGAGC_CCAAAAUCAU ....((((((((...................(((((((..(...)..)))))))..(((......))).............................))))))))............ (-13.45 = -12.48 + -0.97)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:53 2006