| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,494,335 – 13,494,430 |
| Length | 95 |
| Max. P | 0.855725 |

| Location | 13,494,335 – 13,494,430 |
|---|---|
| Length | 95 |
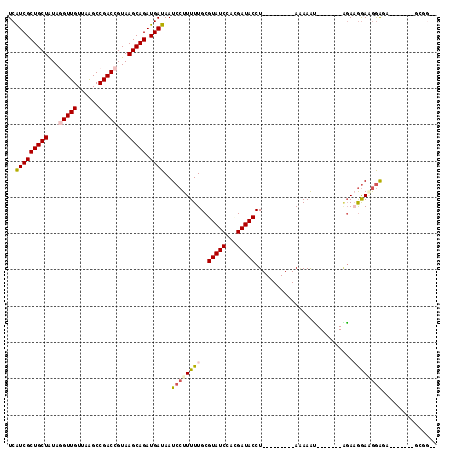
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.31 |
| Mean single sequence MFE | -26.94 |
| Consensus MFE | -18.11 |
| Energy contribution | -18.62 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.66 |
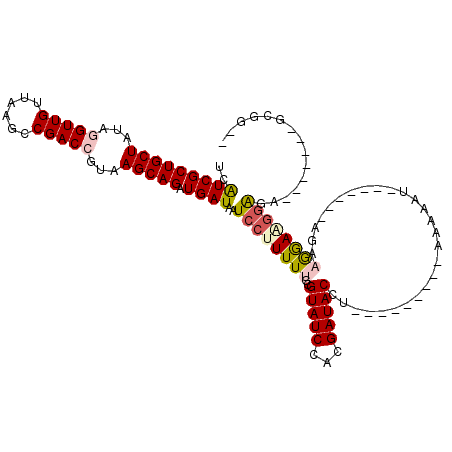
| Structure conservation index | 0.67 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13494335 95 + 20766785 UCAUCGCUGCUGUACGUUGUUAAGCCGACCGUAAGCAGAUGAUAAUCCUUUUUGCGUAUCCACGAUACCU---------AAAAAU-------AGAUGGAUGGGUU-------GCGGGU ..(((((((((.(((((((......))).))))))))).))))...(((....((.(((((((.......---------......-------.).)))))).)).-------..))). ( -23.34) >DroVir_CAF1 4265 103 + 1 UCGUCGCUGCUGUAGGUUGUUAGGCCGACCGUAAGCAGAUGAUAAUCCUUUUUGCGUAUCCAGGAUACCUGCCGAGUACAAAAAA-------UUAAGGAAGAAGA------AGCAG-- ..(((((((((...(((((......)))))...))))).))))..((((((((..((((.((((...))))....))))..))).-------..)))))......------.....-- ( -25.70) >DroGri_CAF1 4532 93 + 1 UCGUCGCUGCUGUAGGUUGUUAAGCCGACCGUAAGCAGAUGAUAAUCCUUUUUGCGUAUCCAGGAUACCU---------AAAAGC-------AGAAGGAAAGAGA------UAAA--- ..(((((((((...(((((......)))))...))))).))))..(((((((.(((((((...)))))..---------....))-------)))))))......------....--- ( -27.20) >DroWil_CAF1 4733 95 + 1 UCAUCGCUGCUAUAGGUUGUUAAGCCGACCGUAAGCAGAUGAUAAUCCUUUUUGCGUAUCCAUGAUACCU---------ACAAACGAAAAGUAGAAAGAGGGAU-------------- ..(((((((((...(((((......)))))...))))).)))).(((((((((..(((((...)))))((---------((.........)))).)))))))))-------------- ( -28.50) >DroAna_CAF1 44498 102 + 1 UCAUCGCUGCUAUAGGUUGUUAAGCCGACCGUAAGCAGAUGAUAAUCCUUUUUGCGUAUCCACGAUACCU---------AAAAAU-------AGAUGAAUGGGUGAGAAAAAGAGGGU ..(((((((((...(((((......)))))...))))).)))).(((((((((...((((((...((.((---------(....)-------)).))..))))))....))))))))) ( -29.50) >DroPer_CAF1 48339 95 + 1 UCGUCGCUGCUAUACGUUGUUAAGCCGACCGUAAGCAGAUGAUAAUCCUUUUUGCGUAUCCAUGAUACCU---------AGAAAU-------GGAAGGAAGGAGA-------ACGGCA ..(((((((((.(((((((......))).))))))))).......((((((((..(((((...)))))((---------(....)-------)).))))))))..-------.)))). ( -27.40) >consensus UCAUCGCUGCUAUAGGUUGUUAAGCCGACCGUAAGCAGAUGAUAAUCCUUUUUGCGUAUCCACGAUACCU_________AAAAAU_______AGAAGGAAGGAGA_______GCGG__ ..(((((((((...(((((......)))))...))))).))))..((((((((..(((((...)))))...........................))))))))............... (-18.11 = -18.62 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:52 2006