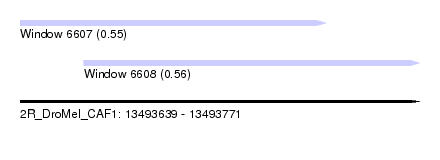
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,493,639 – 13,493,771 |
| Length | 132 |
| Max. P | 0.562690 |

| Location | 13,493,639 – 13,493,740 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.57 |
| Mean single sequence MFE | -32.16 |
| Consensus MFE | -24.42 |
| Energy contribution | -24.75 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.24 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
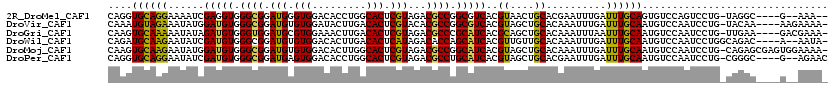
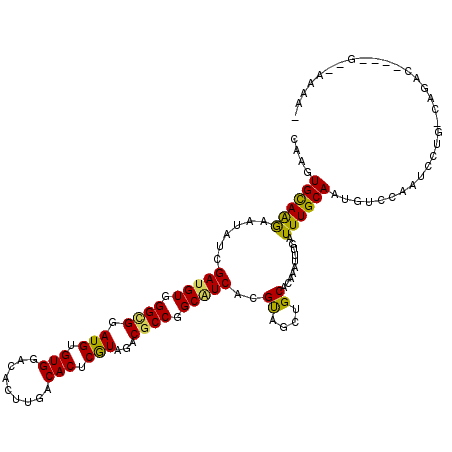
>2R_DroMel_CAF1 13493639 101 + 20766785 ---------------CC----UCUACUUACCGACGACACUCAGGUGCAGGAAAAUCGAGGUGGGCGGAUGGGUGGACACCUGGCACUCGUAGACGCCGGCGUCACGUAACUGCACGAAUU ---------------..----......................((((((.......((.((.((((.(((((((..(....).)))))))...)))).)).))......))))))..... ( -32.32) >DroVir_CAF1 3891 113 + 1 UCGACUGACU---UCUU----UUUACUUACCGACUACGCUCAAAUGUAGAAAUAUGGAUGUGGGCGGAUGUGUGGAUACUUGACACUCGUACACGCCGGCGUCACGUAGCUGCACAAAUU ..........---....----.......................(((((...(((((((((.((((.(((.(((.........))).)))...)))).))))).)))).)))))...... ( -30.40) >DroGri_CAF1 4144 117 + 1 UCAACUAACU---UCUUGCUGUCCACUUACCGACAACACUCAAGUGCAAAAAUAUAGAUGUGGGUGGAUGCGUGGAAACUUGACACUCGUAGACGCCCGCAUCACGCAGCUGCACAAAUU ..........---...((.((((........)))).)).....(((((........((((((((((..((((.(...........).))))..)))))))))).......)))))..... ( -33.86) >DroWil_CAF1 4133 101 + 1 ---------------UC----GUUACUCACCGACAACACUCAGAUGCAAGAAUAUCGAUGUGGGCGGAUGUGUGGACACUUGACACUCAUAGACACCAGCAUCACGUUGUUGCACAAAUU ---------------..----.........(((((((.....((((......))))(((((.((.(.(((.(((.........))).)))...).)).)))))..)))))))........ ( -24.60) >DroMoj_CAF1 4355 116 + 1 UCGGUUGCCCCCGUCGC----UCUACUUACCGACUACACUCAAGUGCAAGAAUAUGGAUGUGGGCGGAUGUGUGGACACUUGGCACUCGUAGACGCCGGCAUCACGUAGCUGCACAAAUU (((((.((.......))----.......)))))..........(((((....(((((((((.((((.(((.(((..(....).))).)))...)))).))))).))))..)))))..... ( -35.90) >DroPer_CAF1 47777 101 + 1 ---------------CC----UCUACUUACCGACGACACUCAGGUGCAGGAAUAUCGAUGUGGGCGGAUGAGUGGACACCUGGCACUCGUAGACGCCUGCAUCACGUAGCUGCACGAAUU ---------------..----......................((((((...(((.((((..((((.(((((((..(....).)))))))...))))..))))..))).))))))..... ( -35.90) >consensus _______________UC____UCUACUUACCGACAACACUCAAGUGCAAGAAUAUCGAUGUGGGCGGAUGUGUGGACACUUGACACUCGUAGACGCCGGCAUCACGUAGCUGCACAAAUU ...........................................((((((.......(((((.((((.(((.(((.........))).)))...)))).)))))......))))))..... (-24.42 = -24.75 + 0.34)


| Location | 13,493,660 – 13,493,771 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -24.51 |
| Energy contribution | -23.73 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13493660 111 + 20766785 CAGGUGCAGGAAAAUCGAGGUGGGCGGAUGGGUGGACACCUGGCACUCGUAGACGCCGGCGUCACGUAACUGCACGAAUUUGAUUUGCAGUGUCCAGUCCUG-UAGGC----G--AAA-- ...((((((.......((.((.((((.(((((((..(....).)))))))...)))).)).))......))))))...((((.(((((((.(......))))-)))))----)--)).-- ( -39.62) >DroVir_CAF1 3924 114 + 1 CAAAUGUAGAAAUAUGGAUGUGGGCGGAUGUGUGGAUACUUGACACUCGUACACGCCGGCGUCACGUAGCUGCACAAAUUUGAUUUGCAAUGUCCAAUCCUG-UACAA----AAGAAAA- (((((((((...(((((((((.((((.(((.(((.........))).)))...)))).))))).)))).)))))....))))........(((.((....))-.))).----.......- ( -31.30) >DroGri_CAF1 4181 114 + 1 CAAGUGCAAAAAUAUAGAUGUGGGUGGAUGCGUGGAAACUUGACACUCGUAGACGCCCGCAUCACGCAGCUGCACAAAUUUAAUUUGCAAUGUCCAAUCCUG-UUGAA----GACGAAA- ....((((((......((((((((((..((((.(...........).))))..))))))))))..((....))..........)))))).((((((((...)-)))..----))))...- ( -34.50) >DroWil_CAF1 4154 113 + 1 CAGAUGCAAGAAUAUCGAUGUGGGCGGAUGUGUGGACACUUGACACUCAUAGACACCAGCAUCACGUUGUUGCACAAAUUUGAUUUGCAAUGUCCAAUCCUGGCAGAC----A--AAUA- ....(((((.(((...(((((.((.(.(((.(((.........))).)))...).)).)))))..))).)))))...(((((.(((((...(......)...))))))----)--))).- ( -28.30) >DroMoj_CAF1 4391 118 + 1 CAAGUGCAAGAAUAUGGAUGUGGGCGGAUGUGUGGACACUUGGCACUCGUAGACGCCGGCAUCACGUAGCUGCACAAAUUUGAUUUGCAAUGUCCAAUCCUG-CAGAGCGAGUGGAAAA- ...(((((....(((((((((.((((.(((.(((..(....).))).)))...)))).))))).))))..)))))...(((.((((((..(((........)-))..)))))).)))..- ( -41.30) >DroPer_CAF1 47798 113 + 1 CAGGUGCAGGAAUAUCGAUGUGGGCGGAUGAGUGGACACCUGGCACUCGUAGACGCCUGCAUCACGUAGCUGCACGAAUUUGAUUUGCAAUGUCCAAUCCUG-CGGGC----G--AGAAC ...((((((...(((.((((..((((.(((((((..(....).)))))))...))))..))))..))).))))))...............(((((.......-.))))----)--..... ( -41.10) >consensus CAAGUGCAAGAAUAUCGAUGUGGGCGGAUGUGUGGACACUUGACACUCGUAGACGCCGGCAUCACGUAGCUGCACAAAUUUGAUUUGCAAUGUCCAAUCCUG_CAGAC____G__AAAA_ ....((((((......(((((.((((.(((.(((.........))).)))...)))).)))))..((....))..........))))))............................... (-24.51 = -23.73 + -0.77)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:51 2006