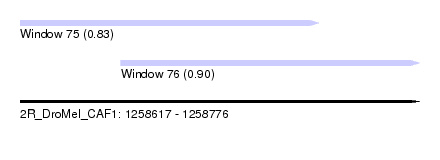
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,258,617 – 1,258,776 |
| Length | 159 |
| Max. P | 0.897915 |

| Location | 1,258,617 – 1,258,736 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 95.80 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -34.70 |
| Energy contribution | -35.18 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
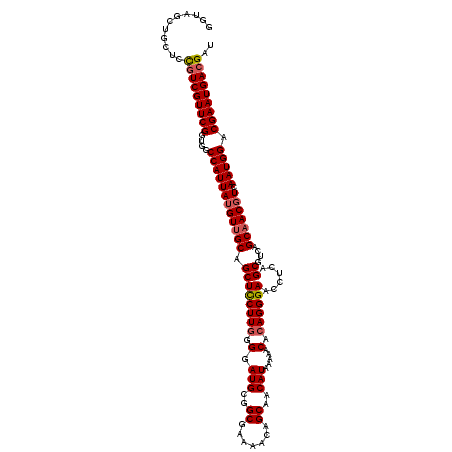
>2R_DroMel_CAF1 1258617 119 + 20766785 GGUAGCUGCUCCGUCGUUCGGUGGCCAUUAUGUUGCAGCUCCUUGGGGAUGCGGCGAAAACAGCAACAUAAAAACACAGGGAACCUCAGCGUCAGCAACGUCAAUGGACGAAUGACGAU ...........(((((((((....((((((((((((.(((((((((..(((..((.......))..))).....).))))))......))....))))))).))))).))))))))).. ( -39.80) >DroSec_CAF1 10170 119 + 1 GGUAGCUGCUCCGUCGUUCGGUGGCCAUUAUGUUGCAGCUCCUUGGGGAUGCGGCGAAAACAGCAACAUAAAAACACAGGGAACCUCAGCGUCAGCAACGUCAAUGGACGAAUGACGAU ...........(((((((((....((((((((((((.(((((((((..(((..((.......))..))).....).))))))......))....))))))).))))).))))))))).. ( -39.80) >DroSim_CAF1 16082 119 + 1 GGUAGCUGCUCCGUCGUUCGGUGGCCAUUAUGUUGCAGCUCCUUGGGGAUGCGGCGAAAACAGCAACAUAAAAACACAGGGAACCUCAGCGUCAGCAACGUCAAUGGACGAAUGACGAU ...........(((((((((....((((((((((((.(((((((((..(((..((.......))..))).....).))))))......))....))))))).))))).))))))))).. ( -39.80) >DroEre_CAF1 14852 119 + 1 GGUAGCUGGUCUGUCGUUCGGUGGCCAUUAUGUUGCAGCUCCUUGGGGAUGCGGCGAAAGCAGCAACAUAAAAACACAGGGAACCUUAGCGUCAGCCACUUCAAUGGACGAAUGACGAU ........(((.(((((((((((((..(((((((((.(((((....))).)).((....)).)))))))))..((.((((....))).).))..))))).((....))))))))))))) ( -42.50) >DroYak_CAF1 13308 119 + 1 GGUAGCUGGUUCUUCGUUCGGUGGCCAUUAUGUUGCAGCUUCUUAGGGAUGCUGCGAAAACAGCAACAUAAAAACACAGGGAACCUCAGCAUCAGCAACGUCAAUGGACGAAUGACGAU ..........((.(((((((....((((((((((((..((....)).((((((((.......))..............(((...))))))))).))))))).))))).))))))).)). ( -33.70) >consensus GGUAGCUGCUCCGUCGUUCGGUGGCCAUUAUGUUGCAGCUCCUUGGGGAUGCGGCGAAAACAGCAACAUAAAAACACAGGGAACCUCAGCGUCAGCAACGUCAAUGGACGAAUGACGAU ...........(((((((((....((((((((((((.((((((((.(.(((..((.......))..))).....).))))))......))....))))))).))))).))))))))).. (-34.70 = -35.18 + 0.48)

| Location | 1,258,657 – 1,258,776 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 96.97 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -26.42 |
| Energy contribution | -26.50 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
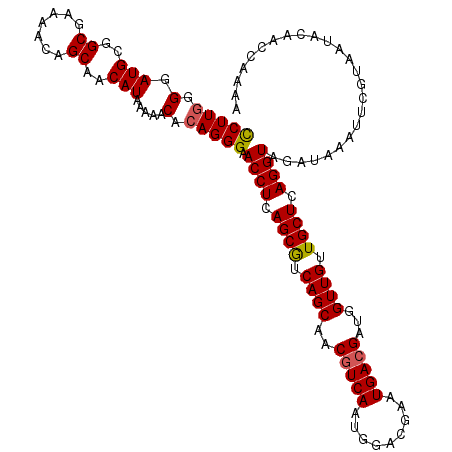
>2R_DroMel_CAF1 1258657 119 + 20766785 CCUUGGGGAUGCGGCGAAAACAGCAACAUAAAAACACAGGGAACCUCAGCGUCAGCAACGUCAAUGGACGAAUGACGAUGGUUGUUGCUCAGGUAGAUAAAUUCGUAAUACAACCAAAA ((((((..(((..((.......))..))).....).)))))........(((((....((((....))))..))))).(((((((....(..((......))..)....)))))))... ( -29.40) >DroSec_CAF1 10210 119 + 1 CCUUGGGGAUGCGGCGAAAACAGCAACAUAAAAACACAGGGAACCUCAGCGUCAGCAACGUCAAUGGACGAAUGACGAUGGUUGUUGCUCAGGUAGAUAAAUUCGUAAUACAACCAAAA ((((((..(((..((.......))..))).....).)))))........(((((....((((....))))..))))).(((((((....(..((......))..)....)))))))... ( -29.40) >DroSim_CAF1 16122 119 + 1 CCUUGGGGAUGCGGCGAAAACAGCAACAUAAAAACACAGGGAACCUCAGCGUCAGCAACGUCAAUGGACGAAUGACGAUGGUUGUUGCUCAGGUAGAUAAAUUCGUAAUACAACCAAAA ((((((..(((..((.......))..))).....).)))))........(((((....((((....))))..))))).(((((((....(..((......))..)....)))))))... ( -29.40) >DroEre_CAF1 14892 119 + 1 CCUUGGGGAUGCGGCGAAAGCAGCAACAUAAAAACACAGGGAACCUUAGCGUCAGCCACUUCAAUGGACGAAUGACGAUGGUUGUUGCUCAGGUAGAUAAAUUCGUAAUACAACCAAAA (((((..(.(((.((....)).))).).........))))).((((.((((.(((((((.(((.........))).).)))))).)))).))))......................... ( -31.30) >DroYak_CAF1 13348 119 + 1 UCUUAGGGAUGCUGCGAAAACAGCAACAUAAAAACACAGGGAACCUCAGCAUCAGCAACGUCAAUGGACGAAUGACGAUGGUUGUUGCUCAGGUAGAUAAAUUCUUAAUACAACCAAAA ..((((((((((((......))))..................((((.((((.((((..(((((.........)))))...)))).)))).))))......))))))))........... ( -29.90) >consensus CCUUGGGGAUGCGGCGAAAACAGCAACAUAAAAACACAGGGAACCUCAGCGUCAGCAACGUCAAUGGACGAAUGACGAUGGUUGUUGCUCAGGUAGAUAAAUUCGUAAUACAACCAAAA (((((.(.(((..((.......))..))).....).))))).((((.((((.((((..(((((.........)))))...)))).)))).))))......................... (-26.42 = -26.50 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:28 2006