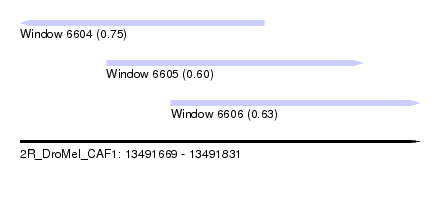
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,491,669 – 13,491,831 |
| Length | 162 |
| Max. P | 0.746617 |

| Location | 13,491,669 – 13,491,768 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -22.83 |
| Energy contribution | -22.22 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13491669 99 - 20766785 CGGCCGCAAGCAUGUGGAUUAAAAAUGCUCGCGCACGGUGCGCCA----------GAU----CCAGCCAUUUGGCAGGCAAACUGCCAUUAA-AGCCAGGCUAUAAACACGGCG- ..(((((.(((((...........))))).))((..(((..((((----------(((----......))))))).(((.....))).....-.)))..)).........))).- ( -32.90) >DroPse_CAF1 38068 113 - 1 CGGCCGCAAGCAUGUGGAUUAAAAAUGCCCGCACACGACACGACGACGGCGACGGCAACACAGCGGCCAUUUGGCAGGCAAACUGCCAUUAA-AGCCAGGCCAUAAACAUACCA- .((((....((.((((.........(((((((...((......))...)))..)))).))))))(((....((((((.....))))))....-.))).))))............- ( -38.80) >DroSec_CAF1 40244 99 - 1 CGGCCGCAAGCAUGUGGAUUAAAAAUGCCCGCGCACGGUGCGCCA----------GAU----CCAGUCAUUUGGCAGGCAAACUGCCAUUAA-AGCCAGGCCAUAAACACGGCG- .((((....((...((((((.....((((((....))).)))...----------)))----)))......((((((.....))))))....-.))..))))............- ( -32.40) >DroYak_CAF1 42197 100 - 1 CGGCCGCAAGCAUGUGGAUUAAAAAUGCCCGCGCACGGUGCUGCA----------GAU----CCAGCCAUUUGGCAGGCAAACUGCCAUUAA-AGCCAGGCCAUAAACACGGCCA .((((....)....((((((......(((((....))).))....----------)))----))))))...((((((.....))))))....-.....((((........)))). ( -34.60) >DroAna_CAF1 42053 100 - 1 CGGCCGCAAGCAUGUGGAUUAAAAAUGCCCGCGCACGGUGUACGG----------CAG----CCAACCAUUUGGCAGGCAAACUGCUAUUAAAAGCCAGCUCAUAAACACAGCG- .........((.((((.........((((((....))).))).((----------(.(----((((....))))).(((.....))).......)))..........)))))).- ( -26.10) >DroPer_CAF1 46116 113 - 1 CGGCCGCAAGCAUGUGGAUUAAAAAUGCCCGCACACGACACGACGACGACGACGGCAACACAGCGGCCAUUUGGCAGGCAAACUGCCAUUAA-AGCCAGGCCAUAAACAUACCA- .((((....((.((((.........((((((....((......))....))..)))).))))))(((....((((((.....))))))....-.))).))))............- ( -34.00) >consensus CGGCCGCAAGCAUGUGGAUUAAAAAUGCCCGCGCACGGUGCGCCA__________CAU____CCAGCCAUUUGGCAGGCAAACUGCCAUUAA_AGCCAGGCCAUAAACACAGCG_ .((((....((.(((((...........)))))......................................((((((.....))))))......))..))))............. (-22.83 = -22.22 + -0.61)


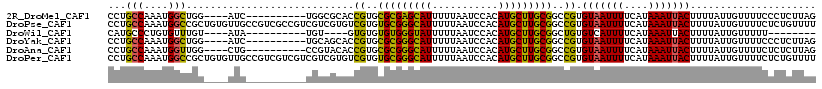
| Location | 13,491,704 – 13,491,808 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.82 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -16.76 |
| Energy contribution | -15.93 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13491704 104 + 20766785 CCUGCCAAAUGGCUGG----AUC----------UGGCGCACCGUGCGCGAGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCCCUCUUAG ((.(((....))).))----...----------.(((((.....))((((((((...........)))))))).)))..(((((((....)))))))..................... ( -28.00) >DroPse_CAF1 38103 118 + 1 CCUGCCAAAUGGCCGCUGUGUUGCCGUCGCCGUCGUCGUGUCGUGUGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCUCUGUUUU ........((((((((.......(((.(((((.(.....).)).))))))((((...........))))..))))))))(((((((....)))))))..................... ( -29.50) >DroWil_CAF1 454 92 + 1 CAUGCCCUGUGUUUGU----AUA----------UGU----GUGUGUGUGGGUAUUUUUAAUCCACAUGCUUGCGGCUGUGUCAUUUUCAUAAAUUACUUUUAUUGUUUUU-------- ((((((..((((....----)))----------)((----(.((((((((((.......)))))))))).)))))).)))..............................-------- ( -17.90) >DroYak_CAF1 42233 104 + 1 CCUGCCAAAUGGCUGG----AUC----------UGCAGCACCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCCCUCUUAG ...(((....))).((----(..----------.((((...((..(((((((((...........)))))))))..)).(((((((....))))))).....))))..)))....... ( -25.60) >DroAna_CAF1 42089 104 + 1 CCUGCCAAAUGGUUGG----CUG----------CCGUACACCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCUCUCUUAG ...(((((....))))----).(----------((((((...))))((((((((...........)))))))))))...(((((((....)))))))..................... ( -25.70) >DroPer_CAF1 46151 118 + 1 CCUGCCAAAUGGCCGCUGUGUUGCCGUCGUCGUCGUCGUGUCGUGUGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCUCUGUUUU ........((((((((......((((.((....)).)).)).(((((.(((.........))).)))))..))))))))(((((((....)))))))..................... ( -28.70) >consensus CCUGCCAAAUGGCUGG____AUG__________CGUCGCACCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCUCUCUUAG ...(((....)))............................((..(((((((((...........)))))))))..)).(((((((....)))))))..................... (-16.76 = -15.93 + -0.83)
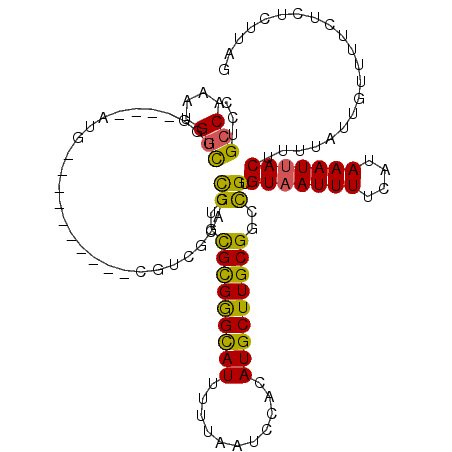
| Location | 13,491,730 – 13,491,831 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -19.17 |
| Consensus MFE | -16.39 |
| Energy contribution | -16.03 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13491730 101 + 20766785 CCGUGCGCGAGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCCCUCUUAGCCAAC---------AGAACCA----ACCACACGCCG--- .((((.((((((((...........))))))))(((...(((((((....))))))).......(........)...)))...---------.......----....))))...--- ( -19.90) >DroPse_CAF1 38143 84 + 1 UCGUGUGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCUCUGUUUUCA------------------------------GUCG--- .((..(((((((((...........)))))))))..)).(((((((....)))))))..............(((....))------------------------------)...--- ( -15.70) >DroEre_CAF1 40669 101 + 1 CCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCCCUCUUAGCCAAC---------AGAGCCA----ACCACGCGCCG--- ..((((((((((((...........))))))))(((...(((((((....))))))).....(((((.............)))---------)).))).----.....))))..--- ( -23.82) >DroYak_CAF1 42259 105 + 1 CCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCCCUCUUAGCCAAC---------AGAACCAACCAACCACACGCCG--- .((((.((((((((...........))))))))(((...(((((((....))))))).......(........)...)))...---------...............))))...--- ( -19.10) >DroAna_CAF1 42115 113 + 1 CCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCUCUCUUAGCAAACGCAUAUAGCAUAACCA----CACUCACUCCACAC ..((((((((((((...........)))))))).((.(((((((((....))))))).....(((((.........))))))))).....)))).....----.............. ( -20.80) >DroPer_CAF1 46191 84 + 1 UCGUGUGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCUCUGUUUUCA------------------------------GUCG--- .((..(((((((((...........)))))))))..)).(((((((....)))))))..............(((....))------------------------------)...--- ( -15.70) >consensus CCGUGCGCGGGCAUUUUUAAUCCACAUGCUUGCGGCCGUGUAAUUUUCAUAAAUUACUUUUAUUGUUUUCCCUCUUAGCAAAC_________AGAACCA____ACCACACGCCG___ .((..(((((((((...........)))))))))..)).(((((((....)))))))............................................................ (-16.39 = -16.03 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:49 2006