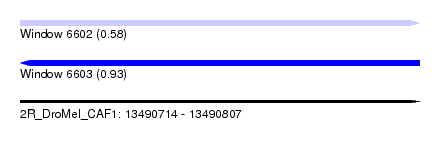
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,490,714 – 13,490,807 |
| Length | 93 |
| Max. P | 0.930277 |

| Location | 13,490,714 – 13,490,807 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 89.45 |
| Mean single sequence MFE | -21.66 |
| Consensus MFE | -16.62 |
| Energy contribution | -16.50 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
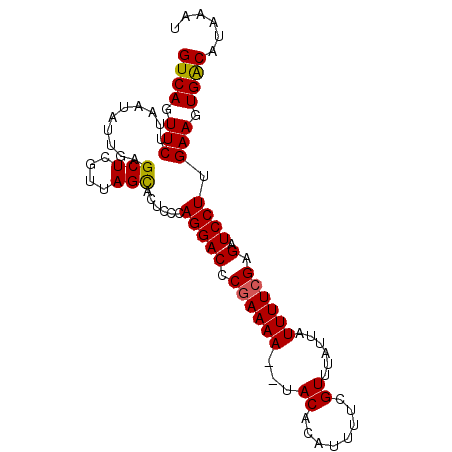
>2R_DroMel_CAF1 13490714 93 + 20766785 GUCAGUUCUUAAUAUUGAGCUCGUUAGCACUACCAGGACCCGAAAA--UACACGUUUUCGUUGAUUAUUUUCGAGAUCCUUGAAGUGACAUAAAU ((((.(((..........(((....)))......((((.(((((((--((.(((....)))....)))))))).).)))).))).))))...... ( -22.30) >DroSec_CAF1 39297 93 + 1 GUCAGUUCUUAAUAUUGAGCUCGUUAGCACUCCCAGGACUCAAAAA--UACGCAAAUUCGUUUAUGAUUUUCGAGAUCCUUGAAGUGGCUUAAAU ...(((((........)))))......((((.(.(((((((.((((--(.((.(((....))).))))))).))).)))).).))))........ ( -18.10) >DroSim_CAF1 39524 93 + 1 GUCAGUUCUUAAUAUUGAGCUCGUUAGCACUCCCAGGACCCGAAAA--UACGCAUUUUCGUUUAUGAUUUUCGAGAUCCUUGAAGUGACAUAAAU ((((.(((..........(((....)))......((((.(((((((--((((......))).....))))))).).)))).))).))))...... ( -19.90) >DroEre_CAF1 39649 93 + 1 GUCAGUUCUUAAUAUUGAGCUCGAUAGCACUGCCAGGACCCGAAAA--UACACGUUUUCGUUACAUAUUUUCGAGGUCCUUGAAGUGACAUGAAU ((((((((........))))).))).(((((.(.((((((((((((--((.(((....)))....)))))))).)))))).).)))).)...... ( -30.50) >DroYak_CAF1 41201 95 + 1 GUCAGUUCUUAAUAUUGAGCUCGUUAGUACUGCCAGGACCCGAAAACUUACUCCUUUUUGUUUAUUAUUUUCGAGAUCCUUGAAGUGACAUGAAU ((((((((........))))).......(((.(.((((.(((((((...((........))......)))))).).)))).).))))))...... ( -17.50) >consensus GUCAGUUCUUAAUAUUGAGCUCGUUAGCACUCCCAGGACCCGAAAA__UACACAUUUUCGUUUAUUAUUUUCGAGAUCCUUGAAGUGACAUAAAU ((((.(((..........(((....)))......(((((.((((((...((........))......)))))).).)))).))).))))...... (-16.62 = -16.50 + -0.12)


| Location | 13,490,714 – 13,490,807 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 89.45 |
| Mean single sequence MFE | -24.90 |
| Consensus MFE | -18.12 |
| Energy contribution | -18.24 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
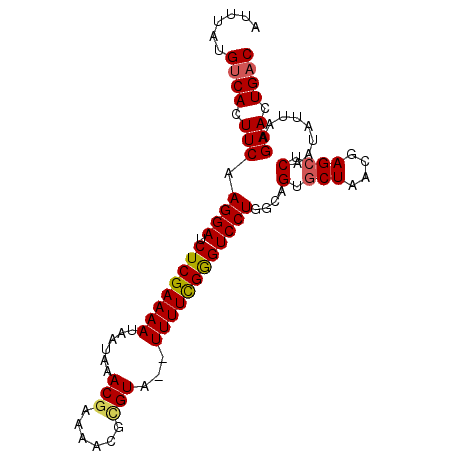
>2R_DroMel_CAF1 13490714 93 - 20766785 AUUUAUGUCACUUCAAGGAUCUCGAAAAUAAUCAACGAAAACGUGUA--UUUUCGGGUCCUGGUAGUGCUAACGAGCUCAAUAUUAAGAACUGAC ......((((.(((.((((((.((((((((....(((....))).))--))))))))))))....(.(((....))).)........))).)))) ( -26.50) >DroSec_CAF1 39297 93 - 1 AUUUAAGCCACUUCAAGGAUCUCGAAAAUCAUAAACGAAUUUGCGUA--UUUUUGAGUCCUGGGAGUGCUAACGAGCUCAAUAUUAAGAACUGAC .....(((.(((((.((((.(((((((((.....(((......))))--)))))))))))).))))))))......................... ( -23.00) >DroSim_CAF1 39524 93 - 1 AUUUAUGUCACUUCAAGGAUCUCGAAAAUCAUAAACGAAAAUGCGUA--UUUUCGGGUCCUGGGAGUGCUAACGAGCUCAAUAUUAAGAACUGAC ......((((.(((.((((((.(((((((.....(((......))))--))))))))))))..((((.(....).))))........))).)))) ( -25.80) >DroEre_CAF1 39649 93 - 1 AUUCAUGUCACUUCAAGGACCUCGAAAAUAUGUAACGAAAACGUGUA--UUUUCGGGUCCUGGCAGUGCUAUCGAGCUCAAUAUUAAGAACUGAC ......((((.(((.((((((.(((((((((...(((....))))))--))))))))))))....(.(((....))).)........))).)))) ( -28.80) >DroYak_CAF1 41201 95 - 1 AUUCAUGUCACUUCAAGGAUCUCGAAAAUAAUAAACAAAAAGGAGUAAGUUUUCGGGUCCUGGCAGUACUAACGAGCUCAAUAUUAAGAACUGAC ......((((.(((.((((((.(((((((......(.....)......))))))))))))).(.(((.(....).))))........))).)))) ( -20.40) >consensus AUUUAUGUCACUUCAAGGAUCUCGAAAAUAAUAAACGAAAACGCGUA__UUUUCGGGUCCUGGCAGUGCUAACGAGCUCAAUAUUAAGAACUGAC ......((((.(((.((((.((((((((......(((......)))...))))))))))))....(.(((....))).)........))).)))) (-18.12 = -18.24 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:47 2006