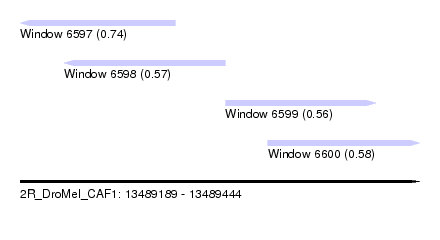
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,489,189 – 13,489,444 |
| Length | 255 |
| Max. P | 0.736836 |

| Location | 13,489,189 – 13,489,288 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 91.21 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -18.35 |
| Energy contribution | -18.10 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13489189 99 - 20766785 ------GCGAAUGUGCCAAUGGCCAAGAGGCAAAUUCCAGUA--GAAGCACAACGGCAACAAAAGUUGGCCACAACAGGCAACAGAUUCUGCAUACGACCAUAAACA ------(.((((.((((..(.....)..)))).))))).(((--(((.(.....(....)....((((.((......)))))).).))))))............... ( -22.10) >DroSec_CAF1 37794 99 - 1 ------GUGAAUGUGCCAAUGGCCAAGAGGCAAAUUCCAGCA--GAAGCACAACGGCAACAAAAGUUGGCCACAACAGGCAACAGAUUCUGCAUACGACCAUAAACA ------(.((((.((((..(.....)..)))).))))).(((--(((.(.....(....)....((((.((......)))))).).))))))............... ( -24.10) >DroEre_CAF1 38138 104 - 1 CGUUUUGCGAAUGUGCCAAUGGCCAAGAGGCAAAUUCCAGCA--GCAACACAACGGCAACAAAAGUUGC-CACAACAGGCAACAGAUUCUGCAUACGACCAUAAACA (((..(((((((.((((....(((....)))...........--..........((((((....)))))-)......))))....)))).))).))).......... ( -28.10) >DroYak_CAF1 39598 105 - 1 --UUUUGUGAAUGUGCCAAUGGCCAAGAGGCAAAUUCCAGCUCGAAAACACAACGGCAACAAAAGUUGCCCACAACAGGCAACAGAUUCUGCAUACGACCAUAAACA --.((((((..((((((....(((....)))...(((......)))........))).)))...((((((.......))))))................)))))).. ( -23.20) >consensus ______GCGAAUGUGCCAAUGGCCAAGAGGCAAAUUCCAGCA__GAAACACAACGGCAACAAAAGUUGCCCACAACAGGCAACAGAUUCUGCAUACGACCAUAAACA ......((((((.((((....(((....))).......................(....)....((((....)))).))))....)))).))............... (-18.35 = -18.10 + -0.25)

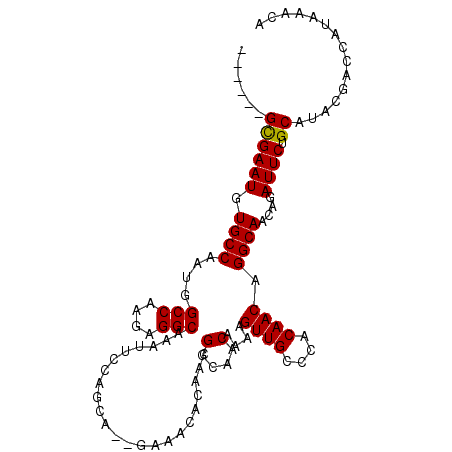
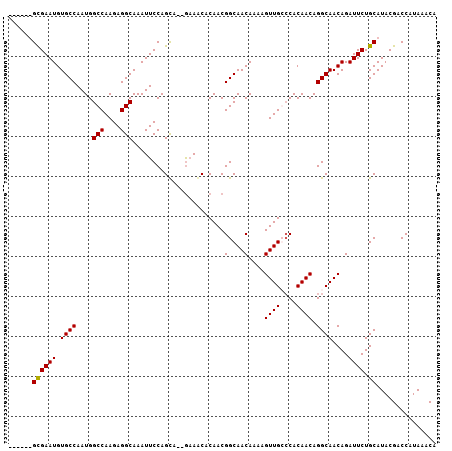
| Location | 13,489,217 – 13,489,320 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 89.60 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -20.35 |
| Energy contribution | -19.75 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13489217 103 - 20766785 AAACCAAAAGCAGAUUGAGCCAAGAGUGUUUU--------------GCGAAUGUGCCAAUGGCCAAGAGGCAAAUUCCAGUA--GAAGCACAACGGCAACAAAAGUUGGCCACAACAGG ...((.............(((....(((((((--------------..((((.((((..(.....)..)))).)))).....--)))))))...))).......((((....)))).)) ( -23.70) >DroSec_CAF1 37822 103 - 1 AAACCAAAAGCAGAUUGAGCCAAGAGUGUUUU--------------GUGAAUGUGCCAAUGGCCAAGAGGCAAAUUCCAGCA--GAAGCACAACGGCAACAAAAGUUGGCCACAACAGG ...((.............(((....(((((((--------------((((((.((((..(.....)..)))).))))..)))--).)))))...))).......((((....)))).)) ( -24.20) >DroSim_CAF1 38039 103 - 1 AAACCAAAAGCAGAUUGAGCCAAGAGUGUUUU--------------GCGAAUGUGCCAAUGGCCAAGAGGCAAAUUCCAGCA--GAAGCACAACGGCAACAAAAGUUGGCCACAACAGG ...((.............(((....(((((((--------------((((((.((((..(.....)..)))).))))..)))--).)))))...))).......((((....)))).)) ( -25.60) >DroEre_CAF1 38166 116 - 1 AAACCAAAAGCAGAUUGAGCCAAGAGUGUUUUGCGUUUUGCGUUUUGCGAAUGUGCCAAUGGCCAAGAGGCAAAUUCCAGCA--GCAACACAACGGCAACAAAAGUUGC-CACAACAGG ...((....................(((((((((..(((((.(((((.......(((...)))))))).))))).....)))--).)))))...((((((....)))))-)......)) ( -30.81) >DroYak_CAF1 39626 112 - 1 AAACCAAAAGCAGAUUGAGCCAAGAGUGUUUUGCG-------UUUUGUGAAUGUGCCAAUGGCCAAGAGGCAAAUUCCAGCUCGAAAACACAACGGCAACAAAAGUUGCCCACAACAGG ...((....................(((((((.((-------..(((.((((.((((..(.....)..)))).)))))))..)))))))))...((((((....)))))).......)) ( -30.00) >consensus AAACCAAAAGCAGAUUGAGCCAAGAGUGUUUU______________GCGAAUGUGCCAAUGGCCAAGAGGCAAAUUCCAGCA__GAAGCACAACGGCAACAAAAGUUGGCCACAACAGG ...((.............(((....(((((((..............((((((.((((..(.....)..)))).))))..))...)))))))...))).......((((....)))).)) (-20.35 = -19.75 + -0.60)

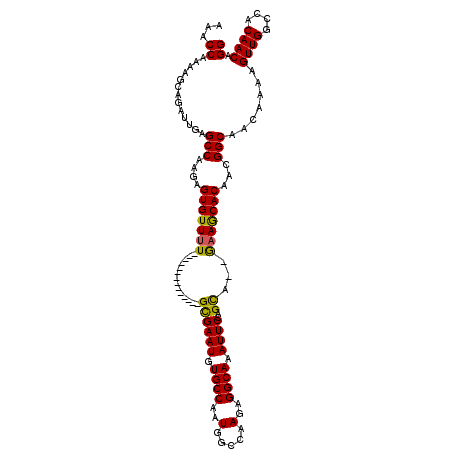
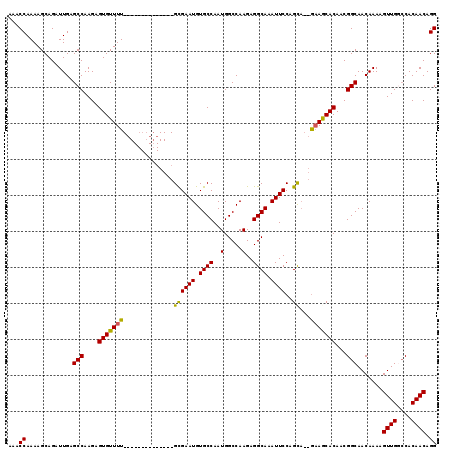
| Location | 13,489,320 – 13,489,416 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -28.79 |
| Consensus MFE | -20.15 |
| Energy contribution | -20.74 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13489320 96 + 20766785 UA-GUACGUCUAUAUUUGCG-UUUU-----------GUUUGCGAGCUUGUGGCUGA--GGAGCUGUGCUCGU-UCGAAUGCGGUUGGUAUAACCAUUAAACUGCAUCGCAAC ..-............(((((-.(((-----------(...((((((..(..(((..--..)))..)))))))-.))))(((((((((.....)))....)))))).))))). ( -28.90) >DroSec_CAF1 37925 107 + 1 UA-GUGCGUCUAUGUUUGCG-UUUUGUUUGCGUUUUGUUUGCGAGCUUGUGGCUGA--GGAGCUGUGCUCGU-UCGAAUGCGGUUGGUAUAACCAUUAAACUGCAUCGCAAC ..-..(((........))).-......(((((.((((...((((((..(..(((..--..)))..)))))))-.))))(((((((((.....)))....)))))).))))). ( -30.90) >DroSim_CAF1 38142 98 + 1 UA-GUGCGUCUAUGUUUGCG-UUUU-----------GUUUGCGAGCUUGUGGCUGAGAGGAGCUGUGCUCGU-UCGAAUGCGGUUGGUAUAACCAUUAAACUGCAUCGCAAC ..-.((((.....(((((((-(((.-----------(...((((((..(..(((......)))..)))))))-.)))))))(((((...)))))...)))).....)))).. ( -29.40) >DroEre_CAF1 38282 97 + 1 UAUGUGUGUCUAUGUUUGCG-UUUU-----------GUUUGCGAGCUUGUGGCUGA--GGAGCUGUGCUCGU-UCGAAUGCGGUUGUUAUAGCCAUUAAACUGCAUCGCAAC ..(((((((....(((((((-....-----------...)))))))..((((((((--(.((((((..((..-..))..)))))).)).)))))))......))).)))).. ( -30.20) >DroYak_CAF1 39738 98 + 1 UAUGUGUGUCUAUGUUUGCG-UUUU-----------GUUUGCGAGCUUGUGGCUGA--GGAGCUGUGCUCGUUUCAAAUGCGGUUGGUAUAAUCAUUAAACUGCAUCGCAAC ...............(((((-.(((-----------(...((((((..(..(((..--..)))..)))))))..))))(((((((.............))))))).))))). ( -28.02) >DroAna_CAF1 39500 95 + 1 UU-CAAUGUCUAUGUUUGCUAUUCUGUGGGAGU------UUUGCGUUUGUGGCUCU--GGAGUUUGGCCCGU-UUGGAUGCGGUUAGCCCAGACAU-----UGCAUCGCA-- ..-((((((((..(((.((((..((.(.(((((------(..........))))))--.)))..))))((((-......))))..)))..))))))-----)).......-- ( -25.30) >consensus UA_GUGCGUCUAUGUUUGCG_UUUU___________GUUUGCGAGCUUGUGGCUGA__GGAGCUGUGCUCGU_UCGAAUGCGGUUGGUAUAACCAUUAAACUGCAUCGCAAC ...............((((.....................((((((....((((......))))..)))))).....((((((((.............)))))))).)))). (-20.15 = -20.74 + 0.59)
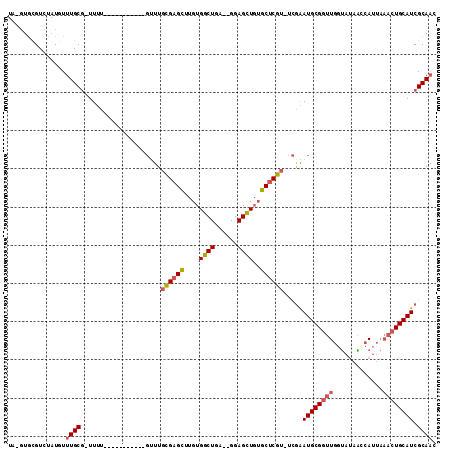
| Location | 13,489,347 – 13,489,444 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.11 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -17.08 |
| Energy contribution | -17.39 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
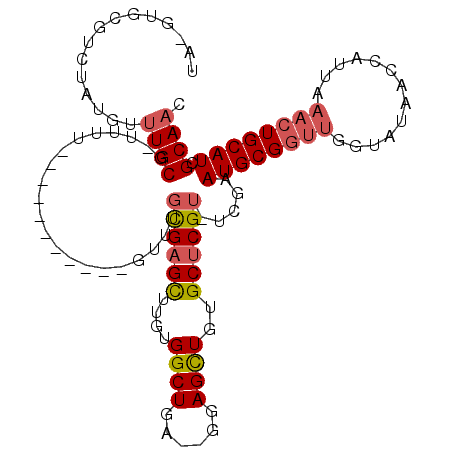
>2R_DroMel_CAF1 13489347 97 + 20766785 GCGAGCUUGUGGCUGA--GGAGCUGUGCUCGU-UCGAAUGCGGUUGGUAUAACCAUUAAACUGCAUCGCAACCGCA------------GUGCUCCUCCAUUAUUUAAGCCAA .........(((((((--(((((..(((..((-((((.(((((((((.....)))....))))))))).))).)))------------..))))))).........))))). ( -36.00) >DroSec_CAF1 37963 97 + 1 GCGAGCUUGUGGCUGA--GGAGCUGUGCUCGU-UCGAAUGCGGUUGGUAUAACCAUUAAACUGCAUCGCAACCGCA------------GCGCUCCUCCAUUAUUUAAGCCAA .........(((((((--(((((..(((..((-((((.(((((((((.....)))....))))))))).))).)))------------..))))))).........))))). ( -35.50) >DroSim_CAF1 38169 99 + 1 GCGAGCUUGUGGCUGAGAGGAGCUGUGCUCGU-UCGAAUGCGGUUGGUAUAACCAUUAAACUGCAUCGCAACCGCA------------GCGCUCCUCCAUUAUUUAAGCCAA ..(.(((((..(.((.(((((((..(((..((-((((.(((((((((.....)))....))))))))).))).)))------------..))))))))).)...)))))).. ( -36.30) >DroEre_CAF1 38310 97 + 1 GCGAGCUUGUGGCUGA--GGAGCUGUGCUCGU-UCGAAUGCGGUUGUUAUAGCCAUUAAACUGCAUCGCAACCGCA------------CUGCUCCUCCAUUAUUUAAGCCAA .........(((((((--(((((.((((..((-((((.(((((((.............)))))))))).))).)))------------).))))))).........))))). ( -37.72) >DroYak_CAF1 39766 110 + 1 GCGAGCUUGUGGCUGA--GGAGCUGUGCUCGUUUCAAAUGCGGUUGGUAUAAUCAUUAAACUGCAUCGCAACCGCAACUGCAACUGCUCCGCUCCUCUAUUAUUUAAGCCAA ..(.((((((((..((--(((((.(.((..(((.((..((((((((......................))))))))..)).))).)).).)))))))..))))..))))).. ( -35.35) >DroAna_CAF1 39533 83 + 1 UUGCGUUUGUGGCUCU--GGAGUUUGGCCCGU-UUGGAUGCGGUUAGCCCAGACAU-----UGCAUCGCA---------------------CACUCCCAUUAUUUAAGGCAA .((((..((..(.(((--((.(((.(((((((-....))).))))))))))))..)-----..)).))))---------------------..................... ( -21.60) >consensus GCGAGCUUGUGGCUGA__GGAGCUGUGCUCGU_UCGAAUGCGGUUGGUAUAACCAUUAAACUGCAUCGCAACCGCA____________GCGCUCCUCCAUUAUUUAAGCCAA ((((((....((((......))))..))))))..(((.(((((((.............))))))))))............................................ (-17.08 = -17.39 + 0.31)

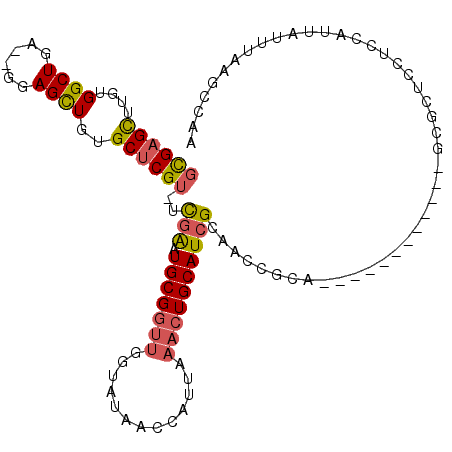
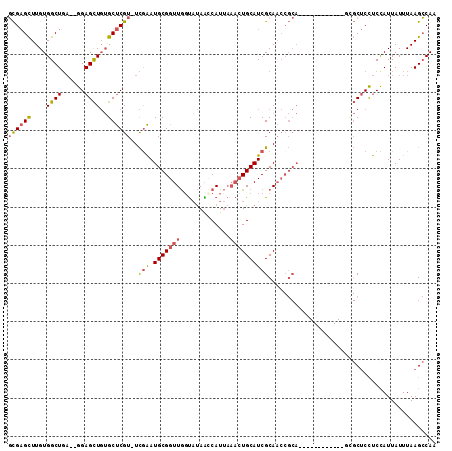
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:44 2006