| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
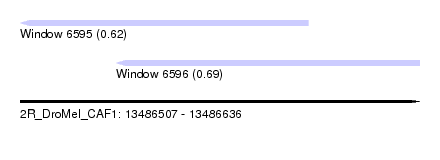
| Location | 13,486,507 – 13,486,636 |
| Length | 129 |
| Max. P | 0.685391 |

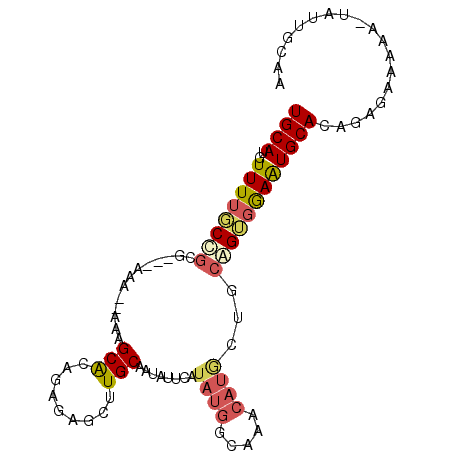
| Location | 13,486,507 – 13,486,600 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 78.28 |
| Mean single sequence MFE | -23.77 |
| Consensus MFE | -8.25 |
| Energy contribution | -8.78 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13486507 93 - 20766785 UGCAUGUUUUGCUGCG---AAAAAAAAGCACAUAGAGCUUGCAAUAUUCAUAUGGUAAACAUGCAGCAGUGGAAUGCACAGAGAAAAA-UAUUGCAA ((((((((((((((((---((....((((.......))))......)))..(((.....)))))))).(((.....))).....))))-)).)))). ( -21.90) >DroSec_CAF1 35055 91 - 1 UGCACGUUUUUCUGCG---AAA--AAAGCAUAGAGAGCUUGCAAUAUUCAUAUGGCAAACAUACUGCAGUGGAAUGCACAGCGAAAAA-UAUUGCAA ((((.((((((((((.---...--...)))......(((((((...(((((...(((.......))).))))).)))).)))))))))-)..)))). ( -22.60) >DroSim_CAF1 35269 91 - 1 UGCAUGUUUUGCUGCG---AAA--AAAGCACAGAGAGCUUGCAAUAUUCAUAUGGCAAACAUCCUGCAGUGGAAUGCACAGCGAAAAA-UAUUGCAA ((((..((((((((..---...--.((((.......))))(((...(((((...(((.......))).))))).))).))))))))..-...)))). ( -22.70) >DroEre_CAF1 35402 91 - 1 UGCAUGUUUGGCCGGG---AAA--AAAGCACAGGGAGCGUGCAAUAUUCAUAUGGCAAACAUGCUGCAGUGGAGUGCACAGGGAAAAA-UAAUUCAA .(((((((((.(((.(---((.--...((((.(....)))))....)))...))))))))))))((((......))))..........-........ ( -25.50) >DroYak_CAF1 36770 92 - 1 UGCAUGUUUUGCCGGGGUAAAA--AGAGCACAGGCAACGUGCAAUAUUCAUAUGGCAAACAUGCAGCAGUGAAAUGCACAGUGAAAAA-U--UGGAA (((((((((.((((........--...((((.(....)))))..........)))))))))))))(((......))).((((.....)-)--))... ( -28.50) >DroAna_CAF1 36798 82 - 1 UGCAUGUUUUUCCGGC---GAA--A--GCGC--------UGCAAUACUCAUAGAGCAAACAAAUGGGGGAGGAAUGCACAGAGAAUAAACUCUAUAA (((((.(((..(((((---...--.--))((--------(.............))).......)))..)))..))))).((((......)))).... ( -21.42) >consensus UGCAUGUUUUGCCGCG___AAA__AAAGCACAGAGAGCUUGCAAUAUUCAUAUGGCAAACAUGCUGCAGUGGAAUGCACAGAGAAAAA_UAUUGCAA ((((..((((((((.............(((.........)))........((((.....))))...))))))))))))................... ( -8.25 = -8.78 + 0.53)


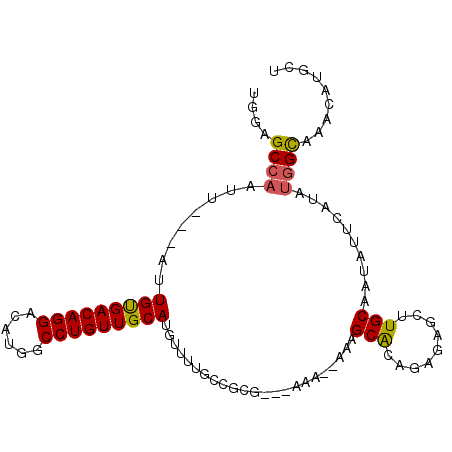
| Location | 13,486,538 – 13,486,636 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 82.76 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -18.83 |
| Energy contribution | -18.75 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13486538 98 - 20766785 UGGAGCCAAUUAUUAUUGUGACAGGACAUGGCCUGUUGCAUGUUUUGCUGCG---AAAAAAAAGCACAUAGAGCUUGCAAUAUUCAUAUGGUAAACAUGCA ....((...(((((((.((((((((......))))(((((.((((((.(((.---........)))..)))))).)))))...)))))))))))....)). ( -22.60) >DroSec_CAF1 35086 96 - 1 UGGAGCCAAUUAUUAUUGUGACAGGACAUGGCCUGUUGCACGUUUUUCUGCG---AAA--AAAGCAUAGAGAGCUUGCAAUAUUCAUAUGGCAAACAUACU ....((((...........((((((......))))))(((.((((((.(((.---...--...)))..)))))).)))..........))))......... ( -24.20) >DroSim_CAF1 35300 96 - 1 UGGAGCCAAUUAUUAUUGUGACAGGACAUGGCCUGUUGCAUGUUUUGCUGCG---AAA--AAAGCACAGAGAGCUUGCAAUAUUCAUAUGGCAAACAUCCU .(((((((.....((((((((((((......))))))((...(((((.(((.---...--...)))))))).))..))))))......)))).....))). ( -25.50) >DroEre_CAF1 35433 93 - 1 UGAAGCCAAUU---AUUGUGACAGGACAUGGCCUGUUGCAUGUUUGGCCGGG---AAA--AAAGCACAGGGAGCGUGCAAUAUUCAUAUGGCAAACAUGCU ...........---.....((((((......))))))(((((((((.(((.(---((.--...((((.(....)))))....)))...)))))))))))). ( -30.30) >DroYak_CAF1 36799 96 - 1 UGAAGCCAAUU---AUUGUGACAGGACAUGGCCUGUUGCAUGUUUUGCCGGGGUAAAA--AGAGCACAGGCAACGUGCAAUAUUCAUAUGGCAAACAUGCA ...........---.....((((((......))))))((((((((.((((........--...((((.(....)))))..........)))))))))))). ( -30.30) >DroAna_CAF1 36830 83 - 1 UGGAGCCAAUU---AUUGCGACAGGACAUGGCCUGUUGCAUGUUUUUCCGGC---GAA--A--GCGC--------UGCAAUACUCAUAGAGCAAACAAAUG ((.(((.....---..(((((((((......)))))))))..........((---...--.--))))--------).))...................... ( -24.20) >consensus UGGAGCCAAUU___AUUGUGACAGGACAUGGCCUGUUGCAUGUUUUGCCGCG___AAA__AAAGCACAGAGAGCUUGCAAUAUUCAUAUGGCAAACAUGCU ....((((........(((((((((......))))))))).......................(((.........)))..........))))......... (-18.83 = -18.75 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:40 2006