| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,485,801 – 13,485,900 |
| Length | 99 |
| Max. P | 0.731153 |

| Location | 13,485,801 – 13,485,900 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.06 |
| Mean single sequence MFE | -19.92 |
| Consensus MFE | -12.21 |
| Energy contribution | -11.77 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
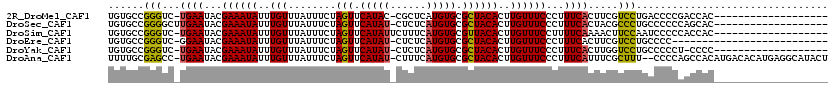
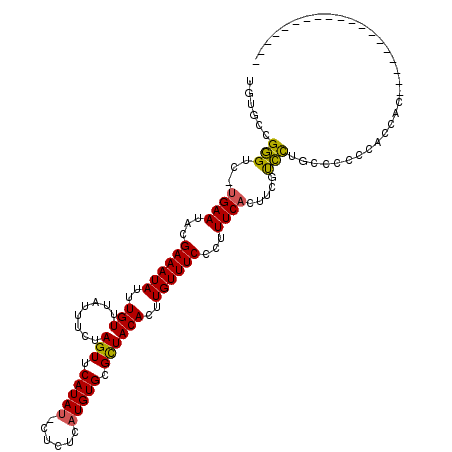
>2R_DroMel_CAF1 13485801 99 + 20766785 UGUGCCGGGUC-UGAAUACGAAAUAUUUGUUUAUUUCUAGUUCAUAC-CGCUCAUGUGCGCUACACUUGUUUCCCUUUCACUUCGUCCUGACCCCGACCAC------------------- ((..(.(((((-(((((..((((((......))))))..)))))...-(((......))).............................))))).)..)).------------------- ( -19.30) >DroSec_CAF1 34410 100 + 1 UGUGCCGGGGCUUGAAUACGAAAUAUUUGUUUAUUUCUAGUUCAUAU-CUCUCAUGUGCGCUACACUUGUUUCCCUUUCACUACGCCCUGCCCCCCAGCAC------------------- .((((.(((((........((((((......))))))((((.(((((-.....))))).))))..........................)))))...))))------------------- ( -23.70) >DroSim_CAF1 34560 100 + 1 UGUGCCGGGUC-UGAAUACGAAAUAUUUGUUUAUUUCUAGUUCAUAUUCUUUCAUGUGCGUUACACUUGUUUCCUUUUCAAAACUUCCAAUCCCCCACCAC------------------- .(((..(((..-(((((..((((((......))))))..)))))........((.(((.....))).))........................)))..)))------------------- ( -13.80) >DroEre_CAF1 34755 92 + 1 UGUGCCGGGUC-GGAAUACGAAAUAUUUGUUUAUUUCUAGUUCAUAU-CUCUCAUGUGCGCUACACUUGUUUCCCUUUCACUUCGUCCUGCCCC-------------------------- ......((((.-(((....((((((......))))))((((.(((((-.....))))).))))......................))).)))).-------------------------- ( -19.50) >DroYak_CAF1 36025 98 + 1 UGUGCCGGGUC-UGAAUACGAAAUAUUUGUUUAUUUCUAGUUCAUAU-CUCUCAUGUGCGCUACACUUGUUUCCCUUUCACUUGGUCCUGCCCCCU-CCCC------------------- ...((.((..(-((((...((((((..(((........(((.(((((-.....))))).))))))..))))))...))))...)..)).)).....-....------------------- ( -18.30) >DroAna_CAF1 36094 116 + 1 UUUUGCGAGCC-UGAAUACGAAAUAUUUGUUUAUUUCUAGUUCAUAU-CUUUCAUGUGCGCUACACUUGUUUCCCUUUCAUUUCGCUUU--CCCCAGCCACAUGACACAUGAGGCAUACU ....(((((..-((((...((((((..(((........(((.(((((-.....))))).))))))..))))))...)))).)))))...--.....(((.((((...)))).)))..... ( -24.90) >consensus UGUGCCGGGUC_UGAAUACGAAAUAUUUGUUUAUUUCUAGUUCAUAU_CUCUCAUGUGCGCUACACUUGUUUCCCUUUCACUUCGUCCUGCCCCCCACCAC___________________ ......(((...((((...((((((..(((........(((.(((((......))))).))))))..))))))...)))).....)))................................ (-12.21 = -11.77 + -0.44)

| Location | 13,485,801 – 13,485,900 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.06 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -13.92 |
| Energy contribution | -14.45 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.731153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
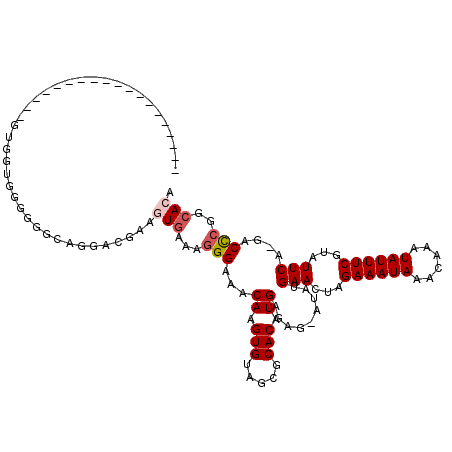
>2R_DroMel_CAF1 13485801 99 - 20766785 -------------------GUGGUCGGGGUCAGGACGAAGUGAAAGGGAAACAAGUGUAGCGCACAUGAGCG-GUAUGAACUAGAAAUAAACAAAUAUUUCGUAUUCA-GACCCGGCACA -------------------(((.(((((......((((((((....(....).((((((.(((......)))-.)))..))).............)))))))).....-..)))))))). ( -28.42) >DroSec_CAF1 34410 100 - 1 -------------------GUGCUGGGGGGCAGGGCGUAGUGAAAGGGAAACAAGUGUAGCGCACAUGAGAG-AUAUGAACUAGAAAUAAACAAAUAUUUCGUAUUCAAGCCCCGGCACA -------------------(((((((((..((..((((..((....(....)...))..))))...))....-...((((...((((((......))))))...))))..))))))))). ( -33.50) >DroSim_CAF1 34560 100 - 1 -------------------GUGGUGGGGGAUUGGAAGUUUUGAAAAGGAAACAAGUGUAACGCACAUGAAAGAAUAUGAACUAGAAAUAAACAAAUAUUUCGUAUUCA-GACCCGGCACA -------------------(((.((((.........(((((......)))))..(((.....)))......(((((((((.((............)).))))))))).-..)))).))). ( -24.40) >DroEre_CAF1 34755 92 - 1 --------------------------GGGGCAGGACGAAGUGAAAGGGAAACAAGUGUAGCGCACAUGAGAG-AUAUGAACUAGAAAUAAACAAAUAUUUCGUAUUCC-GACCCGGCACA --------------------------.(((..((((((((((....(....)..(((.....))).......-......................)))))))...)))-..)))...... ( -19.70) >DroYak_CAF1 36025 98 - 1 -------------------GGGG-AGGGGGCAGGACCAAGUGAAAGGGAAACAAGUGUAGCGCACAUGAGAG-AUAUGAACUAGAAAUAAACAAAUAUUUCGUAUUCA-GACCCGGCACA -------------------....-...((......))..(((...(((...((.(((.....))).))....-...((((...((((((......))))))...))))-..)))..))). ( -21.10) >DroAna_CAF1 36094 116 - 1 AGUAUGCCUCAUGUGUCAUGUGGCUGGGG--AAAGCGAAAUGAAAGGGAAACAAGUGUAGCGCACAUGAAAG-AUAUGAACUAGAAAUAAACAAAUAUUUCGUAUUCA-GGCUCGCAAAA .((..((((((((((((((.(.(((....--..))).).))))...(....)..........)))))))...-...((((...((((((......))))))...))))-)))..)).... ( -29.60) >consensus ___________________GUGGUGGGGGGCAGGACGAAGUGAAAGGGAAACAAGUGUAGCGCACAUGAGAG_AUAUGAACUAGAAAUAAACAAAUAUUUCGUAUUCA_GACCCGGCACA .......................................(((...(((...((.(((.....))).)).........(((...((((((......))))))...)))....)))..))). (-13.92 = -14.45 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:37 2006