| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,482,531 – 13,482,668 |
| Length | 137 |
| Max. P | 0.604994 |

| Location | 13,482,531 – 13,482,634 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 84.45 |
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -16.14 |
| Energy contribution | -17.17 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
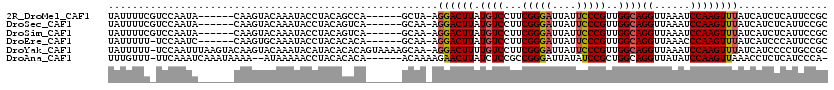
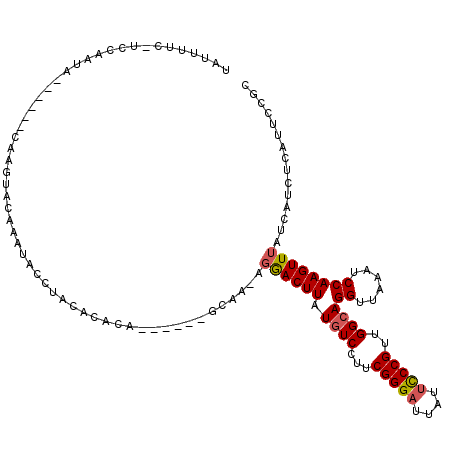
>2R_DroMel_CAF1 13482531 103 + 20766785 -------AAAAGUUGGAAUAAAGAAAAACAA-AAGGCAGCGGAAUGAGAUGAUAAACUUGGAUUUAACCUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCU-UAGC------UGG -------...((((.................-..(((((..(((((((........)))..))))...)))))..(((((....)))))((((((....)))))-))))------).. ( -25.50) >DroSec_CAF1 31198 104 + 1 -------AAAAGUUGGAACAAAGAAAAACAAAAAGGCAGCGGAAUGAGAUGAUAAACUUGGAUUUAACCUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCU-UUGC------UGA -------.............................(((((((..(((........)))(((((((...((((..(((((....)))))..)).))))))))))-))))------)). ( -24.70) >DroSim_CAF1 31346 104 + 1 -------AAAAGUUGGAACAAAGAAAAACAAAAAGGCAGCGGAAUGAGAUGAUAAACUUGGAUUUAACCUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCU-UUGC------UGA -------.............................(((((((..(((........)))(((((((...((((..(((((....)))))..)).))))))))))-))))------)). ( -24.70) >DroEre_CAF1 31571 103 + 1 -------AAAAGUUGGAACAAAGAAAAACAA-AAGGCAGCGGAAUGGGAUGAUAAACUUGGGUUUAACCUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCU-UUGC------UGU -------........................-...(((((.....(((((..(((((....))))).((((....))))....)))))(((((((....)))))-))))------))) ( -29.40) >DroYak_CAF1 32726 116 + 1 AAAAAAAAAAAGUUGGAACAAAGAAAAACAA-AAGGCAGCGGCAGGGGAUGAUAAACUUGGAUUUAACCUGCCAACGGGAAUAAUCCCGAAGGACAAAAGUCCU-UUGCUUUUACUGU ..............................(-((((((..((((((....(((........)))...))))))..(((((....)))))((((((....)))))-))))))))..... ( -32.50) >DroAna_CAF1 32565 103 + 1 -------AAAAGUUAAAAGAAAGAAGACUGGGAAAGC--UGGGAUGAGAGGUUUAACUUGGAUAUAACCUGCCAGCGGAUAUAAUCCCGGCGGAGAUAAGUUCUUUUGU------UGU -------...........((((((..(((......((--((((((..(((......)))...((((.((.......)).)))))))))))).......)))))))))..------... ( -22.62) >consensus _______AAAAGUUGGAACAAAGAAAAACAA_AAGGCAGCGGAAUGAGAUGAUAAACUUGGAUUUAACCUGCCAACGGGAAUAAUCCCGAAGGACAUAAGUCCU_UUGC______UGU ..................................(((((..(((((((........)))..))))...)))))..(((((....)))))..((((....))))............... (-16.14 = -17.17 + 1.03)

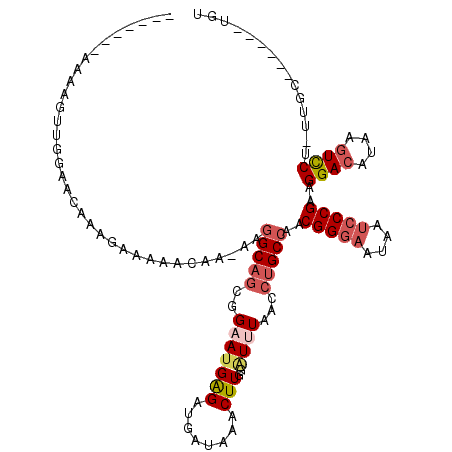
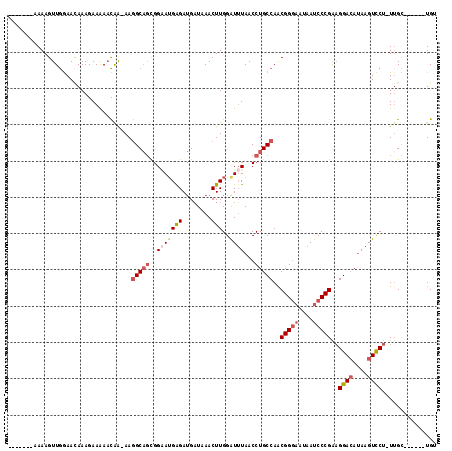
| Location | 13,482,531 – 13,482,634 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 84.45 |
| Mean single sequence MFE | -22.52 |
| Consensus MFE | -14.78 |
| Energy contribution | -15.33 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13482531 103 - 20766785 CCA------GCUA-AGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAGUUUAUCAUCUCAUUCCGCUGCCUU-UUGUUUUUCUUUAUUCCAACUUUU------- (((------((.(-(((((....)))))).((((....))))))))).((((........((........)).........)))).-........................------- ( -20.93) >DroSec_CAF1 31198 104 - 1 UCA------GCAA-AGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAGUUUAUCAUCUCAUUCCGCUGCCUUUUUGUUUUUCUUUGUUCCAACUUUU------- ..(------((((-(((((....))))..(((((....)))))..(((((.............................)))))..))))))...................------- ( -21.55) >DroSim_CAF1 31346 104 - 1 UCA------GCAA-AGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAGUUUAUCAUCUCAUUCCGCUGCCUUUUUGUUUUUCUUUGUUCCAACUUUU------- ..(------((((-(((((....))))..(((((....)))))..(((((.............................)))))..))))))...................------- ( -21.55) >DroEre_CAF1 31571 103 - 1 ACA------GCAA-AGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAACCCAAGUUUAUCAUCCCAUUCCGCUGCCUU-UUGUUUUUCUUUGUUCCAACUUUU------- .((------((.(-(((((....))))))(((((....))))).(((...(.(((((....))))).)...)))....))))....-........................------- ( -24.80) >DroYak_CAF1 32726 116 - 1 ACAGUAAAAGCAA-AGGACUUUUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAGUUUAUCAUCCCCUGCCGCUGCCUU-UUGUUUUUCUUUGUUCCAACUUUUUUUUUUU ((((.((((((((-(((((....))))..(((((....)))))..((((((.(((((....)))))......)))))).......)-))))))))..))))................. ( -30.50) >DroAna_CAF1 32565 103 - 1 ACA------ACAAAAGAACUUAUCUCCGCCGGGAUUAUAUCCGCUGGCAGGUUAUAUCCAAGUUAAACCUCUCAUCCCA--GCUUUCCCAGUCUUCUUUCUUUUAACUUUU------- ...------..(((((((.............((((...))))(((((.(((((............)))))......)))--))..............))))))).......------- ( -15.80) >consensus ACA______GCAA_AGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAGUUUAUCAUCUCAUUCCGCUGCCUU_UUGUUUUUCUUUGUUCCAACUUUU_______ ...............((((....))))..(((((....)))))..(((((.............................))))).................................. (-14.78 = -15.33 + 0.56)

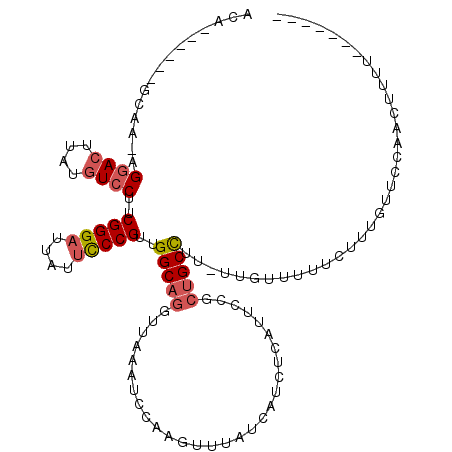
| Location | 13,482,561 – 13,482,668 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.50 |
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -14.19 |
| Energy contribution | -14.42 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13482561 107 - 20766785 UAUUUUCGUCCAAUA------CAAGUACAAAUACCUACAGCCA------GCUA-AGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAGUUUAUCAUCUCAUUCCGC ...............------...........((((...((((------((.(-(((((....)))))).((((....))))))))))))))............................ ( -26.10) >DroSec_CAF1 31229 107 - 1 UAUUUUCGUCCAAUA------CAAGUACAAAUACCUACAGUCA------GCAA-AGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAGUUUAUCAUCUCAUUCCGC ...............------...........((((...((((------((.(-(((((....)))))).((((....))))))))))))))............................ ( -24.00) >DroSim_CAF1 31377 107 - 1 UAUUUUCGUCCAAUA------CAAGUACAAAUACCUACAGUCA------GCAA-AGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAGUUUAUCAUCUCAUUCCGC ...............------...........((((...((((------((.(-(((((....)))))).((((....))))))))))))))............................ ( -24.00) >DroEre_CAF1 31601 106 - 1 UAUUUUU-UCCAAUC------CAAGUGCAAAUACCUACACACA------GCAA-AGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAACCCAAGUUUAUCAUCCCAUUCCGC .......-.......------.....((....((((.....((------((.(-(((((....)))))).((((....))))))))..))))(((((....)))))............)) ( -22.20) >DroYak_CAF1 32763 118 - 1 UAUUUUU-UCCAAUUUAAGUACAAGUACAAAUACAUACACACAGUAAAAGCAA-AGGACUUUUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAGUUUAUCAUCCCCUGCCGC .......-..........(((....))).....................((.(-(((((....))))))(((((....)))))..((((((.(((((....)))))......)))))))) ( -27.00) >DroAna_CAF1 32595 110 - 1 UUUGUUU-UUCAAAUCAAAUAAAA--AUAAAAACCUACACACA------ACAAAAGAACUUAUCUCCGCCGGGAUUAUAUCCGCUGGCAGGUUAUAUCCAAGUUAAACCUCUCAUCCCA- (((((((-((..........))))--)))))(((((.......------.....(((.....)))..((((((........).))))))))))..........................- ( -13.20) >consensus UAUUUUC_UCCAAUA______CAAGUACAAAUACCUACACACA______GCAA_AGGACUUAUGUCCUUCGGGAUUAUUCCCGUUGGCAGGUUAAAUCCAAGUUUAUCAUCUCAUUCCGC .......................................................((((((.((((...(((((....)))))..))))((......))))))))............... (-14.19 = -14.42 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:35 2006