| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,480,815 – 13,480,920 |
| Length | 105 |
| Max. P | 0.582025 |

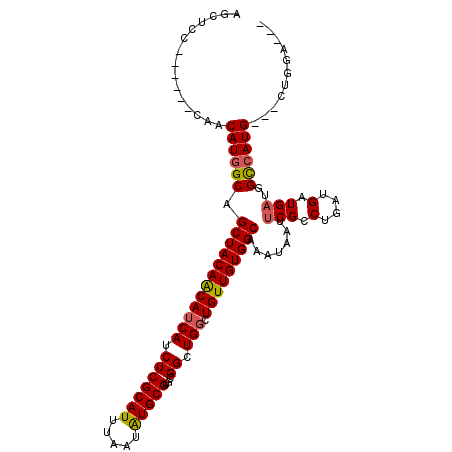
| Location | 13,480,815 – 13,480,920 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 88.52 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -27.72 |
| Energy contribution | -27.80 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13480815 105 + 20766785 AGCUCCAUGGAGCUACAUGGCAGCUACAGCAUCAUCUCGCAUUUAAUAUGCGUGCGGCUGGCUGUUGUGGCAAAUAAUUCGUCUGAUGAUGAUGGCCAUG---CUGGA--- (((((....))))).((((((.((((((((((((.(((((((.....)))))...)).))).))))))))).......(((((....)))))..))))))---.....--- ( -42.40) >DroSec_CAF1 29506 105 + 1 AGCUCCAUGGAGCAACAUGGCAGCUACAACAUCAUCUCGCAUUUAAUAUGCGUGCGGCUGGCUGUUGUGGCAAAUAAUUCGCCUGAUGAUGAUGGCCAUG---CUGGC--- .((((....))))..((((((.((((((((((((.(((((((.....)))))...)).))).))))))))).......(((.(....).)))..))))))---.....--- ( -37.10) >DroSim_CAF1 29635 105 + 1 AGCUCCAUGCAGCAACAUGGCAGCUACAACAUCAUCUCGCAUUUAAUAUGCGUGCGGCUGGCUGUUGUGGCAAAUAAUUCGCCUGAUGAUGAUGGCCAUG---CUGGC--- (((((((((......))))).)))).((.(((((((.(((((.....))))).(((((.....)))))(((.........))).))))))).))(((...---..)))--- ( -35.40) >DroEre_CAF1 29930 98 + 1 AGCUCC-------AACAUGGCAGCUACAACAUCAUCUCGCAUUUAAUGUGCGUGUGGCUGGCUGUUGUGGCAAAUAAUUCGCCUGAUGAUGAUGGCCAUG---CUGGA--- ...(((-------(.((((((.((((((((((((((.(((((.....)))))...)).))).))))))))).......(((.(....).)))..))))))---.))))--- ( -36.20) >DroYak_CAF1 31002 101 + 1 AGCUUC-------AACAUGGCAGCUACAACAUCAUCUCGCAUUUAAUAUGCGUGCGGCUGGCUGUUGUGGCAAAUAAUUCGCCUGAUGAUGAUGGCCAUG---CUGGCGUA .(((..-------..((((((.((((((((((((.(((((((.....)))))...)).))).))))))))).......(((.(....).)))..))))))---..)))... ( -33.50) >DroAna_CAF1 30401 98 + 1 AG----------CAACAUGGCAGCUACAACAUCAUCUCGCAUUUAAUAUGCGUGCGGCUGCCUGUUGUGGCAAAUAAUACGCCUGAUGAUGAUGGUGAUGGCGUUGAA--- .(----------(((((.((((((..((.........(((((.....)))))))..))))))))))))..........(((((................)))))....--- ( -31.59) >consensus AGCUCC______CAACAUGGCAGCUACAACAUCAUCUCGCAUUUAAUAUGCGUGCGGCUGGCUGUUGUGGCAAAUAAUUCGCCUGAUGAUGAUGGCCAUG___CUGGA___ ...............((((((.((((((((((((.(((((((.....)))))...)).))).))))))))).......(((.(....).)))..))))))........... (-27.72 = -27.80 + 0.08)

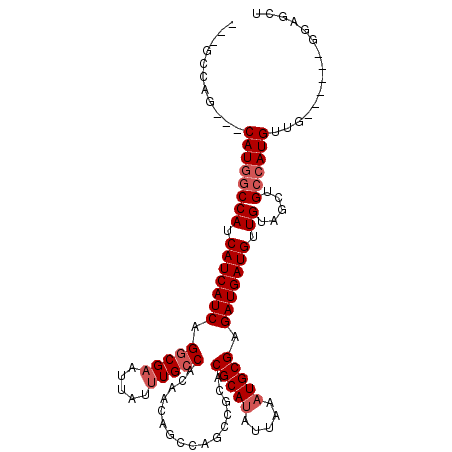
| Location | 13,480,815 – 13,480,920 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 88.52 |
| Mean single sequence MFE | -35.96 |
| Consensus MFE | -25.78 |
| Energy contribution | -26.62 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13480815 105 - 20766785 ---UCCAG---CAUGGCCAUCAUCAUCAGACGAAUUAUUUGCCACAACAGCCAGCCGCACGCAUAUUAAAUGCGAGAUGAUGCUGUAGCUGCCAUGUAGCUCCAUGGAGCU ---....(---((((((((.(((((((......................((.....)).(((((.....))))).))))))).)).....)))))))(((((....))))) ( -33.60) >DroSec_CAF1 29506 105 - 1 ---GCCAG---CAUGGCCAUCAUCAUCAGGCGAAUUAUUUGCCACAACAGCCAGCCGCACGCAUAUUAAAUGCGAGAUGAUGUUGUAGCUGCCAUGUUGCUCCAUGGAGCU ---...((---((((((((.(((((((.(((((.....)))))......((.....)).(((((.....))))).))))))).)).....))))))))((((....)))). ( -39.30) >DroSim_CAF1 29635 105 - 1 ---GCCAG---CAUGGCCAUCAUCAUCAGGCGAAUUAUUUGCCACAACAGCCAGCCGCACGCAUAUUAAAUGCGAGAUGAUGUUGUAGCUGCCAUGUUGCUGCAUGGAGCU ---(((..---...)))((.(((((((.(((((.....)))))......((.....)).(((((.....))))).))))))).)).((((.((((((....)))))))))) ( -38.10) >DroEre_CAF1 29930 98 - 1 ---UCCAG---CAUGGCCAUCAUCAUCAGGCGAAUUAUUUGCCACAACAGCCAGCCACACGCACAUUAAAUGCGAGAUGAUGUUGUAGCUGCCAUGUU-------GGAGCU ---(((((---((((((((.(((((((.(((((.....)))))................((((.......)))).))))))).)).....))))))))-------)))... ( -38.50) >DroYak_CAF1 31002 101 - 1 UACGCCAG---CAUGGCCAUCAUCAUCAGGCGAAUUAUUUGCCACAACAGCCAGCCGCACGCAUAUUAAAUGCGAGAUGAUGUUGUAGCUGCCAUGUU-------GAAGCU ...(((((---((((((((.(((((((.(((((.....)))))......((.....)).(((((.....))))).))))))).)).....))))))))-------)..)). ( -36.60) >DroAna_CAF1 30401 98 - 1 ---UUCAACGCCAUCACCAUCAUCAUCAGGCGUAUUAUUUGCCACAACAGGCAGCCGCACGCAUAUUAAAUGCGAGAUGAUGUUGUAGCUGCCAUGUUG----------CU ---....(((((................)))))...........(((((((((((..(((((((.....))))).........))..)))))).)))))----------.. ( -29.69) >consensus ___GCCAG___CAUGGCCAUCAUCAUCAGGCGAAUUAUUUGCCACAACAGCCAGCCGCACGCAUAUUAAAUGCGAGAUGAUGUUGUAGCUGCCAUGUUG______GGAGCU ...........((((((((.(((((((.(((((.....)))))................(((((.....))))).))))))).)).....))))))............... (-25.78 = -26.62 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:32 2006