| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,462,891 – 13,462,985 |
| Length | 94 |
| Max. P | 0.766436 |

| Location | 13,462,891 – 13,462,985 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.16 |
| Mean single sequence MFE | -34.78 |
| Consensus MFE | -19.60 |
| Energy contribution | -20.80 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
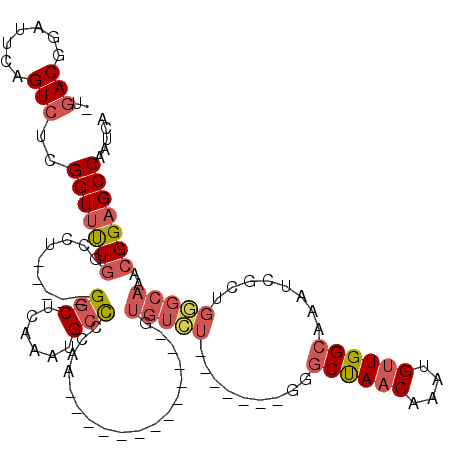
>2R_DroMel_CAF1 13462891 94 + 20766785 -UGACGGAUUCAGUCUCGGUUUUGGCCU-----GGCUCAAAUGCCCCAA-------------GUGUCU-------GGGCUAACAAAUGUUGGCAAAUCGCUGGGCAAACGGAGCCAAUCA -.(((.......)))..(((....)))(-----(((((...(((((..(-------------(((...-------..((((((....))))))....)))))))))....)))))).... ( -35.30) >DroSim_CAF1 11854 94 + 1 -UGACGGAUUCAGUCUCGGUUUUGGCCU-----GGCUCAAAUGCCCCAA-------------GUGUCU-------GGGCUAACAAAUGUUGGCAAAUCGCUGGGCAAACGGAGCCAAUCA -.(((.......)))..(((....)))(-----(((((...(((((..(-------------(((...-------..((((((....))))))....)))))))))....)))))).... ( -35.30) >DroEre_CAF1 12076 94 + 1 -UGACGGAUUCAGUCUCGGUUUUGGCCU-----GGCUCAAAUGCCCCAA-------------GUGUCU-------GGGCUAACAAAUGUUGGCAAAUCGCUGGGCAAACGGAGCCAAUCA -.(((.......)))..(((....)))(-----(((((...(((((..(-------------(((...-------..((((((....))))))....)))))))))....)))))).... ( -35.30) >DroYak_CAF1 12572 94 + 1 -UGACGGAUUCGGUCUCGGUUUUGGCCU-----GGCUCAAAUGCCCCAA-------------GUGUCU-------GGGCUAACAAAUGUUGGCAAAUCGCUGGGCAAACGGAGCCAAUCA -.....((((.((((........)))).-----(((((...(((((..(-------------(((...-------..((((((....))))))....)))))))))....))))))))). ( -36.70) >DroAna_CAF1 7038 120 + 1 CUCACGGAUUCAGUCUAGGUUCUGGCCCGGACCGGCUCGAAUGCCUGACUGUGUGUGCGUCCGUGUGUGGGCGGAGGGCUAACAAAUGUUGGCAAAUCGGUGUGCAAACGGAGCCAAUCA ..(((((..((((((..(((....)))..))).(((......)))))))))))((.((.(((((.((((.((.((..((((((....))))))...)).)).)))).))))))))).... ( -46.60) >DroPer_CAF1 14039 81 + 1 -UGACGGAUUCAGUCUUGGUCUU---CU-----GCCUUCAGUGCUC------------------------------GUCCAACAAAUGAUGGCAAAUCUGUGUGGAAACGGAGCCAAUCU -.(((.......)))((((.(((---(.-----.((..((((((.(------------------------------(((........)))))))...)))...))....))))))))... ( -19.50) >consensus _UGACGGAUUCAGUCUCGGUUUUGGCCU_____GGCUCAAAUGCCCCAA_____________GUGUCU_______GGGCUAACAAAUGUUGGCAAAUCGCUGGGCAAACGGAGCCAAUCA ..(((.......)))..(((((((.........(((......)))..................(((((.........((((((....))))))........)))))..)))))))..... (-19.60 = -20.80 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:27 2006