| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,462,511 – 13,462,616 |
| Length | 105 |
| Max. P | 0.992874 |

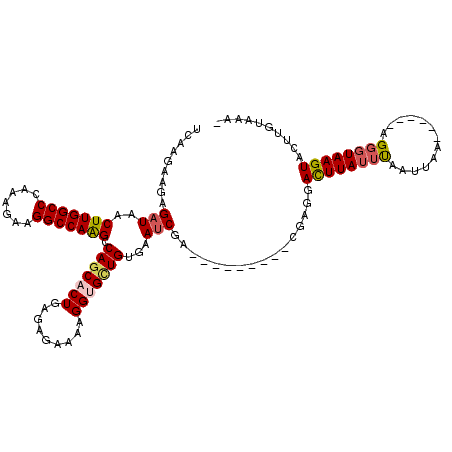
| Location | 13,462,511 – 13,462,616 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.42 |
| Mean single sequence MFE | -24.91 |
| Consensus MFE | -18.70 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13462511 105 + 20766785 CUUUACAAGUACUUAGCCU------UUAAAAAAAUAAGUUCUCC---------UCGAUUCACAGCACCUUUUCUCUCAGUGCUGGCUUGGCCUUCUUUGGGCCAAGUUAUCUCUUCUUGA .....((((.(((((....------.........))))).....---------..(((...((((((...........))))))(((((((((.....))))))))).)))....)))). ( -26.62) >DroSec_CAF1 11303 104 + 1 -UGUACAAGUACUUACACU------UAAAUUAAAUAAAUCCUCG---------UCGAUUCACAGCACCUUUUCUCUCAGUGCUGGCUUGGCCUUCUUUGGGCCAAGUUAUCUCUUCUUGA -....((((..........------...................---------........((((((...........))))))(((((((((.....)))))))))........)))). ( -25.80) >DroSim_CAF1 11461 104 + 1 -UGUACAAGUACUUACCCU------UUAAUUAAAUAAGUCCUCG---------UCGAUUCACAGCACCUUUUCUCUCAGUGCUGGCUUGGCCUUCUUUGGGCCAAGUUAUCUCUUCUUGA -....((((..........------...................---------........((((((...........))))))(((((((((.....)))))))))........)))). ( -25.80) >DroEre_CAF1 11698 97 + 1 ----------ACUUACCCUUCACGUUUAAUCGAAUAAGUCAG------------CGAUUUACAACGCCUUUUCGCUCAGUGAUG-CUUGGCCUCCUUUGGGCCAAGUUAUCUCUUCUUGA ----------............((......))..((((..((------------(((..............))))).((.((((-((((((((.....)))))))).)))).)).)))). ( -22.34) >DroYak_CAF1 12156 107 + 1 -UU------UACUUACCCU------UUAAUAGAAUAAGUCAUCAGGUUGUGGACCGGUUUACAACGCCUUUUCUCUCAGUGCUGGCCUGGCCUUCUUUGGGCCAAGUUAUCUCUUCUUGA -..------.(((((..((------.....))..)))))..((((((((((((....))))))))............((.(.((((.((((((.....)))))).)))).).))..)))) ( -24.00) >consensus _UGUACAAGUACUUACCCU______UUAAUAAAAUAAGUCCUCG_________UCGAUUCACAGCACCUUUUCUCUCAGUGCUGGCUUGGCCUUCUUUGGGCCAAGUUAUCUCUUCUUGA .......................................................(((...((((((...........))))))(((((((((.....))))))))).)))......... (-18.70 = -19.10 + 0.40)

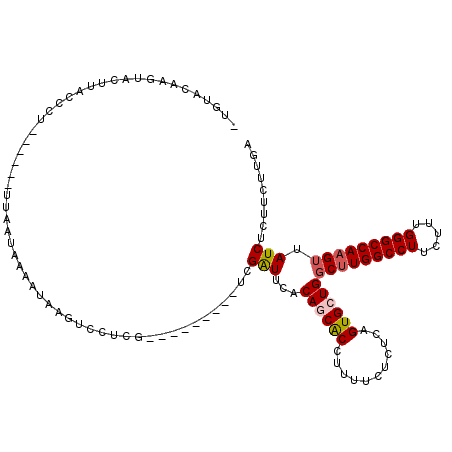
| Location | 13,462,511 – 13,462,616 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.42 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -20.00 |
| Energy contribution | -20.52 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13462511 105 - 20766785 UCAAGAAGAGAUAACUUGGCCCAAAGAAGGCCAAGCCAGCACUGAGAGAAAAGGUGCUGUGAAUCGA---------GGAGAACUUAUUUUUUUAA------AGGCUAAGUACUUGUAAAG ....(((((((((((((((((.......))))))).(((((((.........)))))))........---------.......)))))))))).(------((........)))...... ( -26.00) >DroSec_CAF1 11303 104 - 1 UCAAGAAGAGAUAACUUGGCCCAAAGAAGGCCAAGCCAGCACUGAGAGAAAAGGUGCUGUGAAUCGA---------CGAGGAUUUAUUUAAUUUA------AGUGUAAGUACUUGUACA- .........(((..(((((((.......))))))).(((((((.........)))))))...))).(---------((((.((((((........------...)))))).)))))...- ( -28.00) >DroSim_CAF1 11461 104 - 1 UCAAGAAGAGAUAACUUGGCCCAAAGAAGGCCAAGCCAGCACUGAGAGAAAAGGUGCUGUGAAUCGA---------CGAGGACUUAUUUAAUUAA------AGGGUAAGUACUUGUACA- .........(((..(((((((.......))))))).(((((((.........)))))))...))).(---------((((.((((((((......------.)))))))).)))))...- ( -31.70) >DroEre_CAF1 11698 97 - 1 UCAAGAAGAGAUAACUUGGCCCAAAGGAGGCCAAG-CAUCACUGAGCGAAAAGGCGUUGUAAAUCG------------CUGACUUAUUCGAUUAAACGUGAAGGGUAAGU---------- ......((.(((..((((((((....).)))))))-.))).)).(((((....((...))...)))------------)).((((((((.............))))))))---------- ( -22.92) >DroYak_CAF1 12156 107 - 1 UCAAGAAGAGAUAACUUGGCCCAAAGAAGGCCAGGCCAGCACUGAGAGAAAAGGCGUUGUAAACCGGUCCACAACCUGAUGACUUAUUCUAUUAA------AGGGUAAGUA------AA- (((.......(((((((((((.......))))))(((...............)))))))).....(((.....))))))..(((((((((.....------))))))))).------..- ( -23.66) >consensus UCAAGAAGAGAUAACUUGGCCCAAAGAAGGCCAAGCCAGCACUGAGAGAAAAGGUGCUGUGAAUCGA_________CGAGGACUUAUUUAAUUAA______AGGGUAAGUACUUGUAAA_ .........(((..(((((((.......))))))).(((((((.........)))))))...)))................((((((((.............)))))))).......... (-20.00 = -20.52 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:26 2006