| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,462,305 – 13,462,410 |
| Length | 105 |
| Max. P | 0.890441 |

| Location | 13,462,305 – 13,462,410 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.50 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -26.84 |
| Energy contribution | -27.03 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
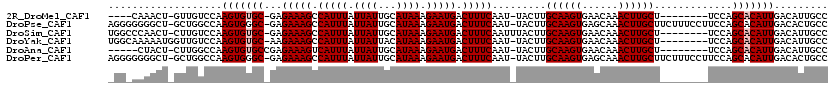
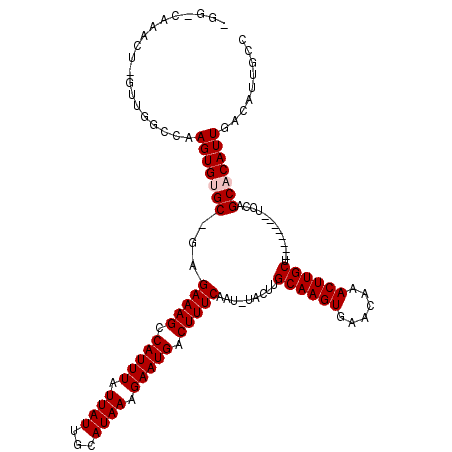
>2R_DroMel_CAF1 13462305 105 + 20766785 GGCAAUGUCAAUGUGCUGGA--------AGCAAGUUUGUUCACUUGCAAGUA-AUUGAAAGUCAUUCUUUAUGCAAUAAUAAAUGGCUUUCUC-GCACACUUGGACAAC-AGUUUG---- .....((((..(((((....--------.((((((......)))))).....-...((((((((((..((((...))))..))))))))))..-)))))....))))..-......---- ( -30.20) >DroPse_CAF1 5983 117 + 1 GGCAGUGUCAAUGUGCUGGAAGGAAAGAAGCAAGUUUGCUCACUUGCAAGUA-AUUGAAAGUCAUUCUUUAUGCAAUAAUAAAUGGCUUUCUC-GCCCACUUGGCCAGC-AGCCCCCCCU (((.((....)).((((((.(((......((((((......)))))).....-...((((((((((..((((...))))..))))))))))..-.....)))..)))))-))))...... ( -33.30) >DroSim_CAF1 11253 110 + 1 GGCAAUGUCAAUGUGCUGGA--------AGCAAGUUUGUUCACUUGCAAGUAAAUUGAAAGUCAUUCUUUAUGCAAUAAUAAAUGGCUUUCUC-GCACACUUGGACAAG-AGUUGGGCCA (((..((((..(((((....--------.((((((......)))))).........((((((((((..((((...))))..))))))))))..-)))))....))))..-......))). ( -32.90) >DroYak_CAF1 11946 110 + 1 GGCAAUGUCAAUGUGCUGGA--------AGCAAGUUUGUUCACUUGCAAGUA-AUUGAAAGUCAUUCUUUAUGUAAUAAUAAAUGGCUUUCUU-GCACACUUGGACAACCAUUUUUGCCA (((((((((..(((((....--------.((((((......)))))).....-...((((((((((..((((...))))..))))))))))..-)))))....)))).......))))). ( -35.11) >DroAna_CAF1 6369 105 + 1 GGCAAUGUCAAUGUGCUGGA--------AGCAAGUUUGUUCACUUGCAAGUA-AUUGAAAGUCAUUCUUUAUGCAAUAAUAAAUGACUUUCUCGGCACACUUGGCCAAG-AGUAG----- ......(((((((((((((.--------.((((((......)))))).....-...((((((((((..((((...))))..)))))))))))))))))).)))))....-.....----- ( -35.10) >DroPer_CAF1 13622 117 + 1 GGCAGUGUCAAUGUGCUGGAAGGAAAGAAGCAAGUUUGCUCACUUGCAAGUA-AUUGAAAGUCAUUCUUUAUGCAAUAAUAAAUGGCUUUCUC-GCCCACUUGGCCAGC-AGCCCCCCCU (((.((....)).((((((.(((......((((((......)))))).....-...((((((((((..((((...))))..))))))))))..-.....)))..)))))-))))...... ( -33.30) >consensus GGCAAUGUCAAUGUGCUGGA________AGCAAGUUUGUUCACUUGCAAGUA_AUUGAAAGUCAUUCUUUAUGCAAUAAUAAAUGGCUUUCUC_GCACACUUGGACAAC_AGUUCC_CC_ .....(((((((((((.............((((((......)))))).........((((((((((..((((...))))..))))))))))...))))).)))).))............. (-26.84 = -27.03 + 0.20)


| Location | 13,462,305 – 13,462,410 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.50 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -22.77 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13462305 105 - 20766785 ----CAAACU-GUUGUCCAAGUGUGC-GAGAAAGCCAUUUAUUAUUGCAUAAAGAAUGACUUUCAAU-UACUUGCAAGUGAACAAACUUGCU--------UCCAGCACAUUGACAUUGCC ----......-..((((..(((((((-..(((((.(((((.((((...)))).))))).)))))...-.....((((((......)))))).--------....)))))))))))..... ( -29.30) >DroPse_CAF1 5983 117 - 1 AGGGGGGGCU-GCUGGCCAAGUGGGC-GAGAAAGCCAUUUAUUAUUGCAUAAAGAAUGACUUUCAAU-UACUUGCAAGUGAGCAAACUUGCUUCUUUCCUUCCAGCACAUUGACACUGCC .(((((((..-(..(((.((((..((-..(((((.(((((.((((...)))).))))).)))))..(-((((....)))))))..))))))).)..))))))).(((.........))). ( -31.60) >DroSim_CAF1 11253 110 - 1 UGGCCCAACU-CUUGUCCAAGUGUGC-GAGAAAGCCAUUUAUUAUUGCAUAAAGAAUGACUUUCAAUUUACUUGCAAGUGAACAAACUUGCU--------UCCAGCACAUUGACAUUGCC .(((......-..((((..(((((((-..(((((.(((((.((((...)))).))))).))))).........((((((......)))))).--------....)))))))))))..))) ( -30.20) >DroYak_CAF1 11946 110 - 1 UGGCAAAAAUGGUUGUCCAAGUGUGC-AAGAAAGCCAUUUAUUAUUACAUAAAGAAUGACUUUCAAU-UACUUGCAAGUGAACAAACUUGCU--------UCCAGCACAUUGACAUUGCC .(((((...(((....)))(((((((-..(((((.(((((.((((...)))).))))).)))))...-.....((((((......)))))).--------....)))))))....))))) ( -33.70) >DroAna_CAF1 6369 105 - 1 -----CUACU-CUUGGCCAAGUGUGCCGAGAAAGUCAUUUAUUAUUGCAUAAAGAAUGACUUUCAAU-UACUUGCAAGUGAACAAACUUGCU--------UCCAGCACAUUGACAUUGCC -----.....-...(((..(((((((.(.(((((((((((.((((...)))).)))))))))))...-.....((((((......)))))).--------..).)))))))......))) ( -32.50) >DroPer_CAF1 13622 117 - 1 AGGGGGGGCU-GCUGGCCAAGUGGGC-GAGAAAGCCAUUUAUUAUUGCAUAAAGAAUGACUUUCAAU-UACUUGCAAGUGAGCAAACUUGCUUCUUUCCUUCCAGCACAUUGACACUGCC .(((((((..-(..(((.((((..((-..(((((.(((((.((((...)))).))))).)))))..(-((((....)))))))..))))))).)..))))))).(((.........))). ( -31.60) >consensus _GG_CAAACU_GUUGGCCAAGUGUGC_GAGAAAGCCAUUUAUUAUUGCAUAAAGAAUGACUUUCAAU_UACUUGCAAGUGAACAAACUUGCU________UCCAGCACAUUGACAUUGCC ...................(((((((...(((((.(((((.((((...)))).))))).))))).........((((((......)))))).............)))))))......... (-22.77 = -23.10 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:24 2006