
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,462,001 – 13,462,209 |
| Length | 208 |
| Max. P | 0.912221 |

| Location | 13,462,001 – 13,462,095 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 88.09 |
| Mean single sequence MFE | -17.62 |
| Consensus MFE | -15.08 |
| Energy contribution | -15.16 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
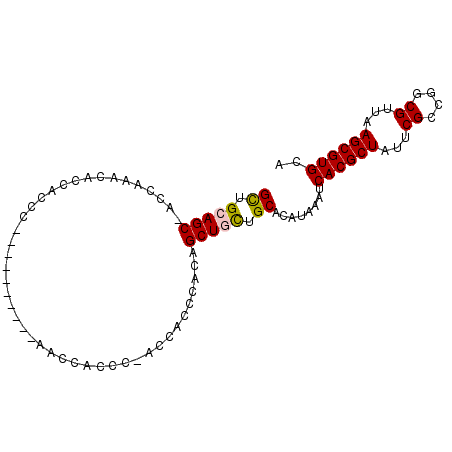
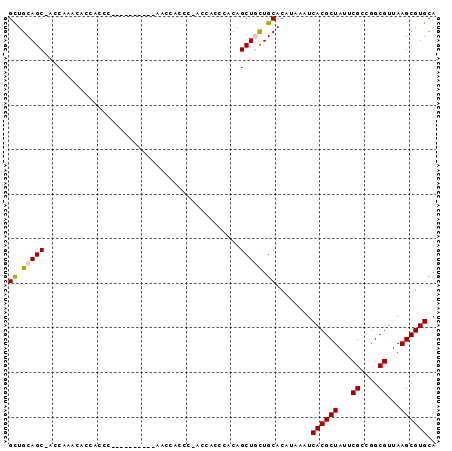
>2R_DroMel_CAF1 13462001 94 - 20766785 GCUGCAGC-ACCAAACACCACCGCCCACCACCCCAACACCC-ACCACCCACAGCUGCUGCACAUAAAUCACGCUAUUCGCCGGCGUUAAGCGUGCA ((.(((((-................................-..........))))).))........((((((...((....))...)))))).. ( -19.05) >DroSec_CAF1 10789 84 - 1 GCUGCAGC-ACCAAACACCACCG----------AACCACCC-ACCACCCACAGCUGCUGCACAUAAAUCACGCUAUUCGCCGGCGUUAAGCGUGCA ((.(((((-..............----------........-..........))))).))........((((((...((....))...)))))).. ( -19.35) >DroSim_CAF1 10957 84 - 1 GCUGCAGC-AGCAAACACCACCA----------AACCACCC-ACCACCCACGGCUGCUGCACAUAAAUCACGCUAUUCGCCGGCGUUAAGCGUGCA ..((((((-(((...........----------........-..........))))))))).......((((((...((....))...)))))).. ( -24.45) >DroEre_CAF1 11187 84 - 1 GCUGCAGCCACCAAACACCACCC----------A-CCACCC-ACCACCCACAGCUAUUGCACAUAAAUCACGCUAUUCGCCGGCGUUAAGCGUGCA (((...(((..............----------.-......-..........((....))...........((.....)).)))....)))..... ( -12.10) >DroYak_CAF1 11655 85 - 1 GUUGCAGCUACCAAACACCAACC----------A-GCACCCAACCACCCACAGCUACUGCACAUAAAUCACGCUAUUCGACGGCGUUAAGCGUGCA ((((..(((..............----------)-))...))))........((....))........((((((...((....))...)))))).. ( -13.14) >consensus GCUGCAGC_ACCAAACACCACCC__________AACCACCC_ACCACCCACAGCUGCUGCACAUAAAUCACGCUAUUCGCCGGCGUUAAGCGUGCA ((.(((((............................................))))).))........((((((...((....))...)))))).. (-15.08 = -15.16 + 0.08)

| Location | 13,462,095 – 13,462,209 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.51 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -20.62 |
| Energy contribution | -21.90 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
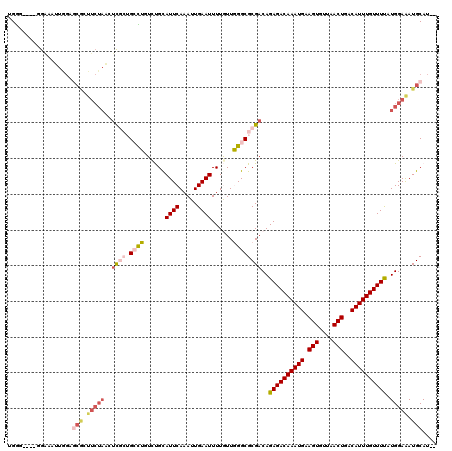
>2R_DroMel_CAF1 13462095 114 - 20766785 UGGG----GGAAAUUGGAGCGCUUCUAACUCGCUGCCUUUUUGCAUUCAAAUUGAAUUUUGUUGGGCGCGACAGAGACAAAUGAAGUGUUAACUGACAUUUGUUUUAUGGAGAUGCAU-- ....----..........(((.(((((..((((.((((......((((.....))))......))))))))..((((((((((.(((....)))..)))))))))).))))).)))..-- ( -31.30) >DroPse_CAF1 5743 119 - 1 UGGUAGAACGGACUUAGUGCACUUCUAGCUCAAAGAUGGUCCGCAUUCAAAUUGAAUUUUAUUUGGCGCGACAGAGACAAAUGAAGUGUUAACUGACAUUUGUUUCAUGGAAAUGCAUC- ................(((((.(((((...........((((((...(((((........)))))))).))).((((((((((.(((....)))..)))))))))).))))).))))).- ( -32.00) >DroEre_CAF1 11271 114 - 1 UGGA----GGAAAUUGGAGCGCUUCUAACUCGCUGCCUUUCUGCAUUCAAAUUGAAUUUUGUUGGGCGCGACAGAGACAAAUGAAGUGUUAACUGACAUUUGUUUUAUGGAGAUGCAU-- ....----..........(((.(((((..((((.((((..(...((((.....))))...)..))))))))..((((((((((.(((....)))..)))))))))).))))).)))..-- ( -32.40) >DroYak_CAF1 11740 114 - 1 GGGG----GGAAAUUGGAACGCUUCUAACUCGCUGCCUUUCUGCAUUCAAAUUGAAUUUUGUUGGGCGCGACAGAGACAAAUGAAGUGUUAACUGACAUUUGUUUUAUGGAGAUGCAU-- ....----............((.(((...((((.((((..(...((((.....))))...)..))))))))((((((((((((.(((....)))..)))))))))).)).))).))..-- ( -30.30) >DroAna_CAF1 6129 106 - 1 GGAGG--AGAAAACAGU------------AUGCUGCUUGUCUGUAUUCAAAUUGAAUUUUGUUGGGCGCGACAGAGACAAAUGAAGUGUUAACUGACAUUUGUUUUAUGGAAAUGCAUCC (((.(--((....(((.------------...)))))).)))((((((...........(((((....)))))((((((((((.(((....)))..))))))))))...).))))).... ( -23.20) >DroPer_CAF1 13382 119 - 1 UGGUAGAACGGACUUAGUGCACUUCUAGCUCAAAGAUGGUCCGCAUUCAAAUUGAAUUUUAUUUGGCGCGACAGAGACAAAUGAAGUGUUAACUGACAUUUGUUUCAUGGAAAUGCAUC- ................(((((.(((((...........((((((...(((((........)))))))).))).((((((((((.(((....)))..)))))))))).))))).))))).- ( -32.00) >consensus UGGG____GGAAAUUGGAGCGCUUCUAACUCGCUGCCUGUCUGCAUUCAAAUUGAAUUUUGUUGGGCGCGACAGAGACAAAUGAAGUGUUAACUGACAUUUGUUUUAUGGAAAUGCAU__ ..................(((.(((((..((((.((((......((((.....))))......))))))))..((((((((((.(((....)))..)))))))))).))))).))).... (-20.62 = -21.90 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:23 2006