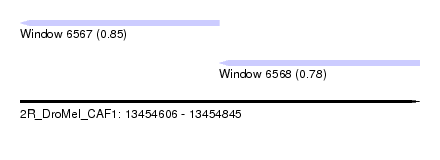
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,454,606 – 13,454,845 |
| Length | 239 |
| Max. P | 0.845306 |

| Location | 13,454,606 – 13,454,725 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.65 |
| Mean single sequence MFE | -26.16 |
| Consensus MFE | -15.07 |
| Energy contribution | -17.03 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
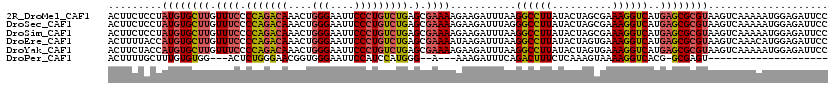
>2R_DroMel_CAF1 13454606 119 - 20766785 CCUUUUUGGCCAUAAA-AAGUUUACCUAUUAGGUGAACUUUCAAAGAAAAUCUCGGGGAGAUAAUUUUUUUUUUUUCAAUCUCAGCACUGCAAAACUUGUGGCACUACAGAUUUUGUUCC ........((((((((-(((((((((.....))))))))))............(((.(((((................)))))....)))......)))))))................. ( -24.79) >DroSec_CAF1 3490 119 - 1 CCUUUUUGGCCAUAAA-AAGUUUACCUAUUAGGUGAACUUUCAAAGAAAAUCUCGGGAAAGUAUUUUUCUUUUUUUCAAUCUCAGCGCUGCAAAAUUUGUGGCACUACAGAUUUUGUUCC .......((((....(-(((((((((.....)))))))))).((((((((...(......)...))))))))............).)))((((((((((((....))))))))))))... ( -28.80) >DroSim_CAF1 3652 119 - 1 CCUUUUUGGCCAUAAA-AAGUUUACCUAUUAGGUGAACUUUCAAAGAAAAUCUCGGGAAAGUAUUUUUCUUUUUUUCAAUCUCAACGCUGCAAAAUUUGUGGCAAUACAGAUUCUGUUCC ........((((((((-(((((((((.....)))))))))..((((((((...(......)...)))))))).......................))))))))...((((...))))... ( -25.50) >DroEre_CAF1 3961 104 - 1 CCUU-UUGGCCAUAAAAAAGUUUACCUAUUAGGUGAACUUCCCAAGAAAAUCUGUGGCAAGUAUUUU----U---UCAAUCUUAGCUUUACAAAA--------ACAGCAAAUUUUGUUUC ....-...(((((....(((((((((.....)))))))))....((.....))))))).........----.---..............((((((--------........))))))... ( -18.20) >DroYak_CAF1 4102 116 - 1 CCUUUUUGGCCAUAAAAAAGUUUACCUAUUAGGUGAACUUUCCAACAAAAUCUGUGGCAAGUAUUUU----UUGUUCAAUCUCAGCGCUACUAAAUUUGUAGCACCACAGAUUUUCUUCC .....((((.......((((((((((.....)))))))))))))).(((((((((((((((.....)----)))............(((((.......))))).)))))))))))..... ( -33.51) >consensus CCUUUUUGGCCAUAAA_AAGUUUACCUAUUAGGUGAACUUUCAAAGAAAAUCUCGGGAAAGUAUUUUU_UUUUUUUCAAUCUCAGCGCUGCAAAAUUUGUGGCACUACAGAUUUUGUUCC ((...............(((((((((.....)))))))))...............))................................((((((((((((....))))))))))))... (-15.07 = -17.03 + 1.96)


| Location | 13,454,725 – 13,454,845 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -22.92 |
| Energy contribution | -23.53 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
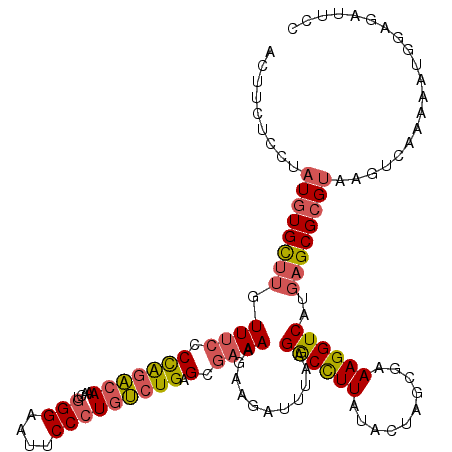
>2R_DroMel_CAF1 13454725 120 - 20766785 ACUUCUCCUAUGUGCUUGUUUCCCCAGACAAACUGGGAAUUCCCUGUCUGAGCGAAAAGAAGAUUUAAGGCCUUAUACUAGCGAAAGGUCAUGAGCGCGUAAGUCAAAAAUGGAGAUUCC ...((((((((((((((.(((((.((((((....(((....))))))))).).))))...........((((((..........))))))..)))))))))..........))))).... ( -35.70) >DroSec_CAF1 3609 120 - 1 ACUUCUCCUAUGUGCUUGUUUCCCCAGACAAACUGGGAAUUCCCUGUCUGAGCGAAAAGAAGAUUUAGGGCCUUAUACUAGCGAAAGGUCAUGAGCGCGUAAGUCAAAAAUGGAGAUUCC ...((((((((((((((.(((((.((((((....(((....))))))))).).))))...........((((((..........))))))..)))))))))..........))))).... ( -35.50) >DroSim_CAF1 3771 120 - 1 ACUUCUCCUAUGUGCUUGUUUCCCCAGACAAACUGGGAAUUCCCUGUCUGAGCGAAAAGAAGAUUUAAGGCCUUAUACUAGCGAAAGGUCAUGAGCGCGUAAGUCAAAAAUGGAGAUUCC ...((((((((((((((.(((((.((((((....(((....))))))))).).))))...........((((((..........))))))..)))))))))..........))))).... ( -35.70) >DroEre_CAF1 4065 120 - 1 ACUUUUACCAUGUGCUUGUUUCCCCAGACAAACUGGGAAUUCCCUGUCUGAGCGAAAAUAAGAUUUAAGGCCUUAUACUAGUGAAAGGUCAUGAGCGCGUAAGUCAAACAUGGAGAUUCC .......((((((((((.(((((.((((((....(((....))))))))).).))))...........((((((.((....)).))))))..))))((....))...))))))....... ( -34.80) >DroYak_CAF1 4218 120 - 1 ACUUCUACCAUGUGCUUGUUUCCCCAGACAAACUGGGAAUUCCCUGUCUGAGCGAAAAGAAGAUUUAAGGCCUUAUACUAGUGAAAGGUCAUGAGCGCGUAAGUCAAAAAUGGAGAUUCC ...(((.((((((((((.(((((.((((((....(((....))))))))).).))))...........((((((.((....)).))))))..))))))(.....)....))))))).... ( -33.10) >DroPer_CAF1 3336 91 - 1 ACUUUUGCUUUGUGUGG---ACUCUGGGAACGGUGGGAAUUCCAUCCAUGGG--A---AAAGAUUUCAGACUUUCUCAAAGUAAAAGGUCACG-GCGAGU-------------------- .((((((((((..((((---((.(((....))).)((....)).)))))(((--(---((...........))))))))))))))))((....-))....-------------------- ( -24.30) >consensus ACUUCUCCUAUGUGCUUGUUUCCCCAGACAAACUGGGAAUUCCCUGUCUGAGCGAAAAGAAGAUUUAAGGCCUUAUACUAGCGAAAGGUCAUGAGCGCGUAAGUCAAAAAUGGAGAUUCC .........((((((((.((((.(((((((....(((....))))))))).).))))...........((((((..........))))))..)))))))).................... (-22.92 = -23.53 + 0.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:14 2006