
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,454,368 – 13,454,473 |
| Length | 105 |
| Max. P | 0.599247 |

| Location | 13,454,368 – 13,454,473 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.32 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -15.82 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
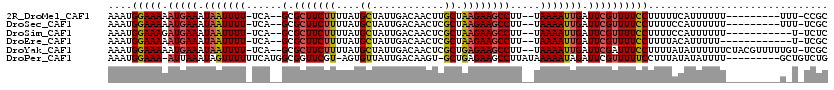
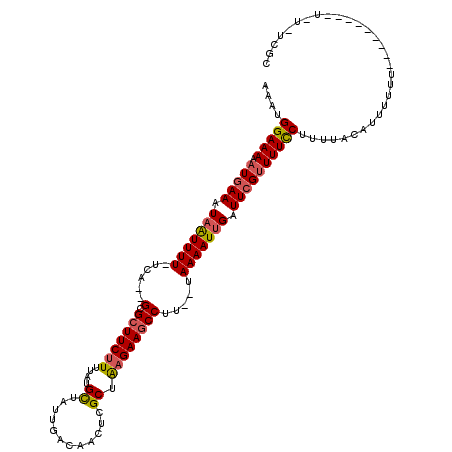
>2R_DroMel_CAF1 13454368 105 - 20766785 AAAUGGAAAAAUGAAAUAAUUUU-UCA--GCGCUUCUUUUAUGCUAUUGACAACUUGCUAAGAAGCCUU--UAAAAUUGAUUCGUUUUCCUUUUUCAUUUUUU---------UUU-CCGC ....((((((.((((.(((((((-..(--(.(((((((....((............)).))))))))).--.))))))).)))))))))).............---------...-.... ( -23.50) >DroSec_CAF1 3253 105 - 1 AAAUGGAAAAAUGAAAUAAUUUU-UCA--GCGCUUCUUUUAUGCUAUUGACAACUCGCUAAGAAGCCUU--UAAAAUUGAUUCGUUUUCCUUUUCCAUUUUUU---------UUU-UCGC (((((((((((((((.(((((((-..(--(.(((((((....((............)).))))))))).--.))))))).))))).....))))))))))...---------...-.... ( -25.10) >DroSim_CAF1 3416 103 - 1 AAAUGGAAAGAUGAAAUAAUUUU-UCA--GCGCUUCUUUUAUGCUAUUGACAACUCGCUAAGAAGCCUU--UAAAAUUGAUUCGUUUUCCUUUUCCAUUUUUU-----------U-UCUC (((((((((((((((.(((((((-..(--(.(((((((....((............)).))))))))).--.))))))).)))))).....)))))))))...-----------.-.... ( -25.50) >DroEre_CAF1 3725 102 - 1 AAAUGGAAAAAUGAAAUAAUUUU-UCA--GCGCUUCUUUUAUGCUAUUGACAACUCGCUAAGAAGCCUU--UAAAAUUGAUUCGUUUUCCUUUUACAUUUUU------------U-UCGC ....((((((.((((.(((((((-..(--(.(((((((....((............)).))))))))).--.))))))).))))))))))............------------.-.... ( -22.90) >DroYak_CAF1 3853 114 - 1 AAAUGGAAAAAUGAAAUAAUUUU-UCA--GCGCUUCUUUUAUGCUAUUGACAACUCGCUGAGAAGCCUU--UAAAAUUGAUUCGAUUUCCUUUUAUAUUUUUUCUACGUUUUUGU-UCGC ....(((((..((((.(((((((-..(--(.(((((((....((............)).))))))))).--.))))))).)))).))))).........................-.... ( -20.90) >DroPer_CAF1 2957 108 - 1 AAAUGGAAA-AUUAAAUAGUUUUUUCAUGGCGGUUCGU-AGUGUUAUUGACAAGU-GCUGAGAAGCCUUAUAAAAAUAGAUUCGUUUUUCCUUUAUAUAUUUU---------GCUGUCUG ....(((((-(...(((.((((((....(((..(((((-(.((((...))))..)-)).)))..)))....))))))..)))...))))))............---------........ ( -18.30) >consensus AAAUGGAAAAAUGAAAUAAUUUU_UCA__GCGCUUCUUUUAUGCUAUUGACAACUCGCUAAGAAGCCUU__UAAAAUUGAUUCGUUUUCCUUUUACAUUUUUU_________U_U_UCGC ....(((((.(((((.(((((((......(.(((((((....((............)).)))))))).....))))))).)))))))))).............................. (-15.82 = -16.02 + 0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:12 2006