
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,452,670 – 13,452,791 |
| Length | 121 |
| Max. P | 0.995300 |

| Location | 13,452,670 – 13,452,770 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.59 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -21.13 |
| Energy contribution | -20.88 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13452670 100 - 20766785 AAUUACUUUCACGUCAGUCAUAUGCACUCGGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUUCCCUCAGC-------------CGAUAGGC--GCACACACA----AAACGA- ............((.(.((((((((((.(((....))))))))))))).).))......................((-------------(....)))--.........----......- ( -27.50) >DroSec_CAF1 1390 100 - 1 AAUUACUUUCACGUCAGUCAUAUGCACUCGGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUUCCCUCAUC-------------CGAUAGGC--GCACACACA----AAACGA- ...........(((((.((((((((((.(((....))))))))))))).)..........(((....))).......-------------.....)))--)........----......- ( -24.40) >DroSim_CAF1 1540 100 - 1 AAUUACUUUCACGUCAGUCAUAUGCACUCGGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUUCCCUCAGC-------------CGAUAGGC--GCACACACA----AAACGA- ............((.(.((((((((((.(((....))))))))))))).).))......................((-------------(....)))--.........----......- ( -27.50) >DroEre_CAF1 2064 100 - 1 AAUUACUUUCACGUCAGUCAUAUGCACUCGGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUUCCCUCAGC-------------CGAUAGGC--GCACACACA----AAACGA- ............((.(.((((((((((.(((....))))))))))))).).))......................((-------------(....)))--.........----......- ( -27.50) >DroYak_CAF1 2182 105 - 1 AAUUACUUUCACGUCAGUCAUAUGCACUCGGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUUCCCUCAGCCGC----------CGAUAGGCGCGCACACACA----AAACGA- ............((.(.((((((((((.(((....))))))))))))).).))......................(((((----------(....)))).)).......----......- ( -32.80) >DroPer_CAF1 1332 120 - 1 AAUUACUUUUACGUCAGUCAUAUGCAAUAGGGUACCUUGUGCGUAUGAGUAACUUAAUUAGAAAACUUUCCCUUGCUGGCAUGAAAAUACAUACAUGCACCCACACACAUAUAAAACCAC .....((.(((.((.(.(((((((((.((((....))))))))))))).).)).)))..)).................(((((..........)))))...................... ( -21.80) >consensus AAUUACUUUCACGUCAGUCAUAUGCACUCGGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUUCCCUCAGC_____________CGAUAGGC__GCACACACA____AAACGA_ ............((.(.((((((((((.(((....))))))))))))).).))................................................................... (-21.13 = -20.88 + -0.25)

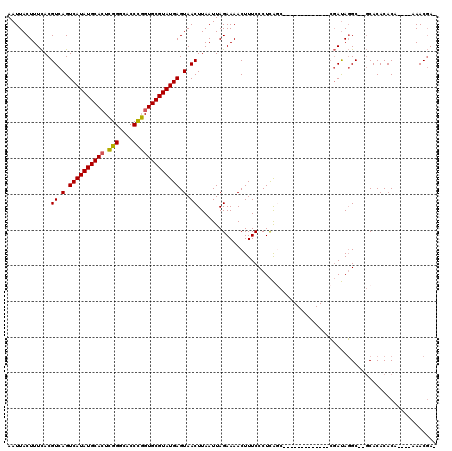
| Location | 13,452,693 – 13,452,791 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.02 |
| Mean single sequence MFE | -30.15 |
| Consensus MFE | -25.98 |
| Energy contribution | -26.90 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13452693 98 + 20766785 ---GCUGAGGGAAAGUUUUCUAAUUAAGUUACUCAUACGCACCGGGUGCCCGAGUGCAUAUGACUGACGUGAAAGUAAUUUUCAUAA--AAGGGAACUUG-----------------GUG ---.........(((((..((......((((.(((((.(((((((....))).)))).))))).))))(((((((...)))))))..--.))..))))).-----------------... ( -31.40) >DroSec_CAF1 1413 99 + 1 ---GAUGAGGGAAAGUUUUCUAAUUAAGUUACUCAUACGCACCGGGUGCCCGAGUGCAUAUGACUGACGUGAAAGUAAUUUUCAUAA--AAGGGAACUUGG----------------GUG ---.........(((((..((......((((.(((((.(((((((....))).)))).))))).))))(((((((...)))))))..--.))..)))))..----------------... ( -31.40) >DroSim_CAF1 1563 99 + 1 ---GCUGAGGGAAAGUUUUCUAAUUAAGUUACUCAUACGCACCGGGUGCCCGAGUGCAUAUGACUGACGUGAAAGUAAUUUUCAUAA--AAGGGAACUUGG----------------GUG ---.........(((((..((......((((.(((((.(((((((....))).)))).))))).))))(((((((...)))))))..--.))..)))))..----------------... ( -31.40) >DroEre_CAF1 2087 98 + 1 ---GCUGAGGGAAAGUUUUCUAAUUAAGUUACUCAUACGCACCGGGUGCCCGAGUGCAUAUGACUGACGUGAAAGUAAUUUUCAUAA--AAGGGAACUUGG-----------------UG ---.........(((((..((......((((.(((((.(((((((....))).)))).))))).))))(((((((...)))))))..--.))..)))))..-----------------.. ( -31.40) >DroYak_CAF1 2207 104 + 1 GCGGCUGAGGGAAAGUUUUCUAAUUAAGUUACUCAUACGCACCGGGUGCCCGAGUGCAUAUGACUGACGUGAAAGUAAUUUUCAUAA--AAGGCAACUUGGUG--------------GUG ((.((..((((((....))))......((((.(((((.(((((((....))).)))).))))).))))(((((((...)))))))..--.......))..)).--------------)). ( -32.60) >DroPer_CAF1 1372 120 + 1 GCCAGCAAGGGAAAGUUUUCUAAUUAAGUUACUCAUACGCACAAGGUACCCUAUUGCAUAUGACUGACGUAAAAGUAAUUUUCAUAUCAACCAUAACUCAGAGGAGAGAGGAAAAGGGUG .((.....))(((((((..((......((((.(((((.(((..(((...)))..))).))))).)))).....)).)))))))......(((....(((........)))......))). ( -22.70) >consensus ___GCUGAGGGAAAGUUUUCUAAUUAAGUUACUCAUACGCACCGGGUGCCCGAGUGCAUAUGACUGACGUGAAAGUAAUUUUCAUAA__AAGGGAACUUGG________________GUG ............(((((((((......((((.(((((.(((((((....))).)))).))))).))))(((((((...))))))).....)))))))))..................... (-25.98 = -26.90 + 0.92)

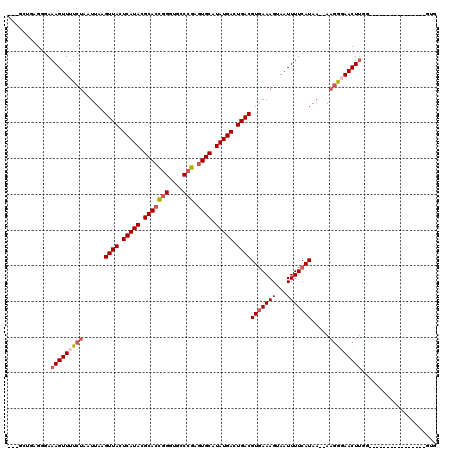
| Location | 13,452,693 – 13,452,791 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.02 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -24.09 |
| Energy contribution | -23.57 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13452693 98 - 20766785 CAC-----------------CAAGUUCCCUU--UUAUGAAAAUUACUUUCACGUCAGUCAUAUGCACUCGGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUUCCCUCAGC--- ...-----------------.(((((..((.--((((((((.....))))).((.(.((((((((((.(((....))))))))))))).).)).)))..))..))))).........--- ( -25.70) >DroSec_CAF1 1413 99 - 1 CAC----------------CCAAGUUCCCUU--UUAUGAAAAUUACUUUCACGUCAGUCAUAUGCACUCGGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUUCCCUCAUC--- ...----------------..(((((..((.--((((((((.....))))).((.(.((((((((((.(((....))))))))))))).).)).)))..))..))))).........--- ( -25.70) >DroSim_CAF1 1563 99 - 1 CAC----------------CCAAGUUCCCUU--UUAUGAAAAUUACUUUCACGUCAGUCAUAUGCACUCGGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUUCCCUCAGC--- ...----------------..(((((..((.--((((((((.....))))).((.(.((((((((((.(((....))))))))))))).).)).)))..))..))))).........--- ( -25.70) >DroEre_CAF1 2087 98 - 1 CA-----------------CCAAGUUCCCUU--UUAUGAAAAUUACUUUCACGUCAGUCAUAUGCACUCGGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUUCCCUCAGC--- ..-----------------..(((((..((.--((((((((.....))))).((.(.((((((((((.(((....))))))))))))).).)).)))..))..))))).........--- ( -25.70) >DroYak_CAF1 2207 104 - 1 CAC--------------CACCAAGUUGCCUU--UUAUGAAAAUUACUUUCACGUCAGUCAUAUGCACUCGGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUUCCCUCAGCCGC ...--------------....(((((((...--...(((((.....)))))......((((((((((.(((....))))))))))))))))))))......................... ( -28.40) >DroPer_CAF1 1372 120 - 1 CACCCUUUUCCUCUCUCCUCUGAGUUAUGGUUGAUAUGAAAAUUACUUUUACGUCAGUCAUAUGCAAUAGGGUACCUUGUGCGUAUGAGUAACUUAAUUAGAAAACUUUCCCUUGCUGGC .............(((....((((((((..((((..(((((.....)))))..))))(((((((((.((((....)))))))))))))))))))))...)))........((.....)). ( -25.90) >consensus CAC________________CCAAGUUCCCUU__UUAUGAAAAUUACUUUCACGUCAGUCAUAUGCACUCGGGCACCCGGUGCGUAUGAGUAACUUAAUUAGAAAACUUUCCCUCAGC___ .....................(((((..........(((((.....))))).((.(.((((((((((.(((....))))))))))))).).))..........)))))............ (-24.09 = -23.57 + -0.53)

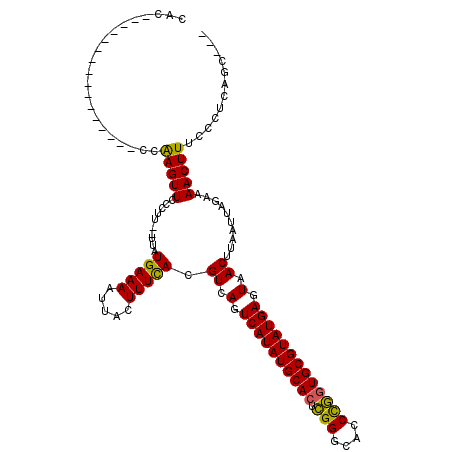
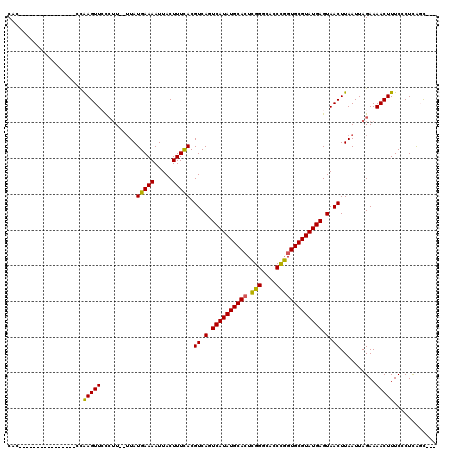
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:09 2006