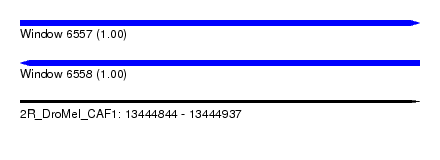
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,444,844 – 13,444,937 |
| Length | 93 |
| Max. P | 0.999834 |

| Location | 13,444,844 – 13,444,937 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 89.76 |
| Mean single sequence MFE | -19.50 |
| Consensus MFE | -17.94 |
| Energy contribution | -17.94 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.20 |
| SVM RNA-class probability | 0.999834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
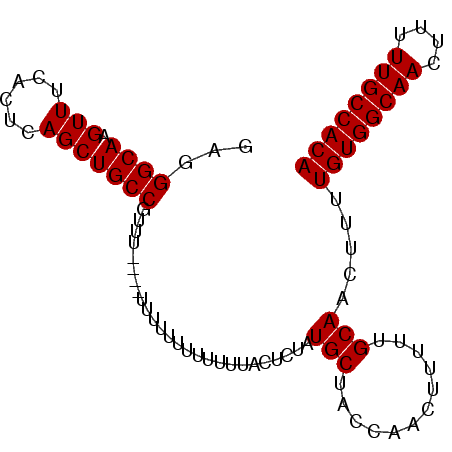
>2R_DroMel_CAF1 13444844 93 + 20766785 GCGGGCAAGUUUCACUCAGCUGCCGUUUUGUUUUUUUUUUUUUUACUCUAUGCUACCAACUUUUUGCAACUUUUGUGGCAACUUUUUGCCACA ...((((.(((......)))))))..........................(((............))).....((((((((....)))))))) ( -18.40) >DroSec_CAF1 8762 90 + 1 GAGGGCAAGUUUCACUCAGCUGCCGCUUC---UUUUUUUUUUUUACUCUAUGCUACCAACUUUUUGCAACUUUUGUGGCAACUUUUUGCCACA (((((((.(((......))))))).))).---..................(((............))).....((((((((....)))))))) ( -19.20) >DroSim_CAF1 9414 87 + 1 GAGGGCAAGUUUCACUCAGCUGCCGUU------UUUUUUUUUUUACUCUAUGCUACCAACUUUUUGCAACUUUUGUGGCAACUUUUUGCCACA ((.((((.(((......))))))).))------.................(((............))).....((((((((....)))))))) ( -18.60) >DroEre_CAF1 8840 82 + 1 GAGGGCAAGUUUCACUCAGCUGCCUUU-----------UUUUUUACUCUAUGCCACCAACUUUUUGCAACUUUUGUGGCAACUUUUUGCCACA (((((((.(((......))))))))))-----------............(((............))).....((((((((....)))))))) ( -19.10) >DroYak_CAF1 8879 93 + 1 GCGGGCAAGUUUCACUCAGCUGCCUGUUCUUUUUUUUUAUUUUUACGCUGUGCUACCAACUUUUUGCAACUUUUGUGGCAACUUUUUGCCACA (((((((.(((......)))))))))).......................(((............))).....((((((((....)))))))) ( -22.20) >consensus GAGGGCAAGUUUCACUCAGCUGCCGUUU____UUUUUUUUUUUUACUCUAUGCUACCAACUUUUUGCAACUUUUGUGGCAACUUUUUGCCACA ...((((.(((......)))))))..........................(((............))).....((((((((....)))))))) (-17.94 = -17.94 + -0.00)

| Location | 13,444,844 – 13,444,937 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 89.76 |
| Mean single sequence MFE | -20.04 |
| Consensus MFE | -18.74 |
| Energy contribution | -18.58 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.93 |
| SVM RNA-class probability | 0.999710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
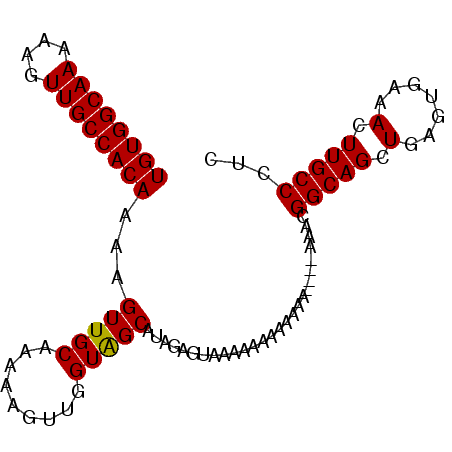
>2R_DroMel_CAF1 13444844 93 - 20766785 UGUGGCAAAAAGUUGCCACAAAAGUUGCAAAAAGUUGGUAGCAUAGAGUAAAAAAAAAAAAAACAAAACGGCAGCUGAGUGAAACUUGCCCGC ((((((((....))))))))...(((((.........)))))..........................(((..((.(((.....)))))))). ( -19.10) >DroSec_CAF1 8762 90 - 1 UGUGGCAAAAAGUUGCCACAAAAGUUGCAAAAAGUUGGUAGCAUAGAGUAAAAAAAAAAAA---GAAGCGGCAGCUGAGUGAAACUUGCCCUC ((((((((....)))))))).(((((.((...(((((...((...................---...))..)))))...)).)))))...... ( -21.15) >DroSim_CAF1 9414 87 - 1 UGUGGCAAAAAGUUGCCACAAAAGUUGCAAAAAGUUGGUAGCAUAGAGUAAAAAAAAAAA------AACGGCAGCUGAGUGAAACUUGCCCUC ((((((((....))))))))...(((((.........)))))..................------...(((((.(.......).)))))... ( -18.90) >DroEre_CAF1 8840 82 - 1 UGUGGCAAAAAGUUGCCACAAAAGUUGCAAAAAGUUGGUGGCAUAGAGUAAAAAA-----------AAAGGCAGCUGAGUGAAACUUGCCCUC ((((((((....)))))))).(((((.((...(((((.(.((.....))......-----------...).)))))...)).)))))...... ( -19.50) >DroYak_CAF1 8879 93 - 1 UGUGGCAAAAAGUUGCCACAAAAGUUGCAAAAAGUUGGUAGCACAGCGUAAAAAUAAAAAAAAAGAACAGGCAGCUGAGUGAAACUUGCCCGC .(.((((...((((..(((...((((((.....((((......))))((.................))..))))))..))).))))))))).. ( -21.53) >consensus UGUGGCAAAAAGUUGCCACAAAAGUUGCAAAAAGUUGGUAGCAUAGAGUAAAAAAAAAAAA____AAACGGCAGCUGAGUGAAACUUGCCCUC ((((((((....))))))))...(((((.........)))))...........................(((((.(.......).)))))... (-18.74 = -18.58 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:04 2006