
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,430,050 – 13,430,176 |
| Length | 126 |
| Max. P | 0.846297 |

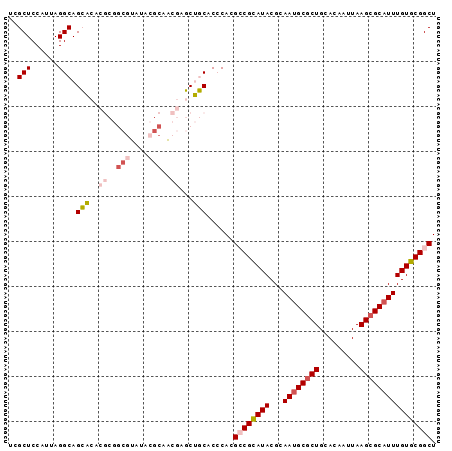
| Location | 13,430,050 – 13,430,145 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 83.47 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -25.83 |
| Energy contribution | -26.80 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13430050 95 + 20766785 UCGCUCCAUUAGGCAGCACACGCGGCGUAUACGCAACGAGCUGCACCCACGCCGCAUACGCAAUGCGCUGCACAAUUAAGCGCAUUUGUGCGGCU ............(((((...((..(((....)))..)).)))))......((((((((...((((((((.........)))))))))))))))). ( -39.90) >DroPse_CAF1 27390 86 + 1 UUGCUCCAUUAGGCAGGC---------UCUAUUCAGGGUUCGUCGCACACGGCGUAUACGGAAUGCGCUGCACAAUUAAGCGCAUUUGUGCGGCU (((((......)))))((---------(((....)))))..(((((((.((.......))(((((((((.........)))))))))))))))). ( -31.90) >DroSec_CAF1 27328 95 + 1 CCGCUCCAUUAGGCAGCACACGCGGCGUAUACGCAACGAGCUGCACCCACGCCGCAUACGCAAUGCGCUGCACAAUUAAGCGCAUUUGUGCGGCU ............(((((...((..(((....)))..)).)))))......((((((((...((((((((.........)))))))))))))))). ( -39.90) >DroSim_CAF1 28621 95 + 1 UCGCUCCAUUAGGCAGCACACGCGGCGUAUACGCAACGAGCUGCACCCACGCCGCAUACGCAAUGCGCUGCACAAUUAAGCGCUUUUGUGCGGCU ............(((((...((..(((....)))..)).)))))......((((((((......(((((.........)))))...)))))))). ( -36.10) >DroYak_CAF1 28640 95 + 1 UCGCUCCAUUAGGCAGCACACGCGGCGUAUACGCAACGAGCUGCACCCACGCCGCAUACGCAAUGCGCUGCACAAUUAAGCGCAUUUGUGCGGCU ............(((((...((..(((....)))..)).)))))......((((((((...((((((((.........)))))))))))))))). ( -39.90) >DroPer_CAF1 27485 84 + 1 UUGCUCCAUUAGGCAGGC---------UCUAU--GGGGUUCGUCGCACACGGCGUAUACGGAAUGCACUGCACAAUUAAGCGCAUUUGUGCGGCU ..(((((((..((....)---------)..))--)))))..(((((((.((.......))((((((.((.........)).))))))))))))). ( -30.10) >consensus UCGCUCCAUUAGGCAGCACACGCGGCGUAUACGCAACGAGCUGCACCCACGCCGCAUACGCAAUGCGCUGCACAAUUAAGCGCAUUUGUGCGGCU ..(((......))).(((..((..(((....)))..))...)))......((((((((...((((((((.........)))))))))))))))). (-25.83 = -26.80 + 0.97)


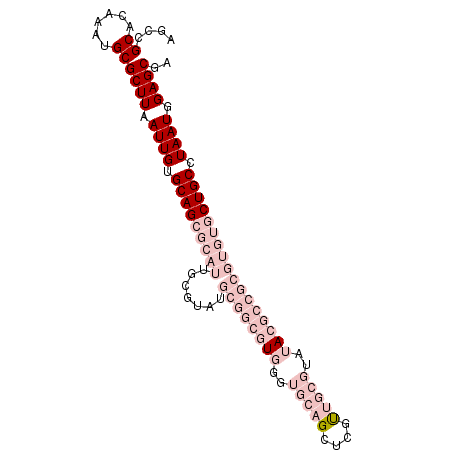
| Location | 13,430,050 – 13,430,145 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 83.47 |
| Mean single sequence MFE | -37.79 |
| Consensus MFE | -26.42 |
| Energy contribution | -31.53 |
| Covariance contribution | 5.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13430050 95 - 20766785 AGCCGCACAAAUGCGCUUAAUUGUGCAGCGCAUUGCGUAUGCGGCGUGGGUGCAGCUCGUUGCGUAUACGCCGCGUGUGCUGCCUAAUGGAGCGA ....(((....)))((((.((((.(((((((((.......((((((((..(((((....)))))..))))))))))))))))).)))).)))).. ( -44.91) >DroPse_CAF1 27390 86 - 1 AGCCGCACAAAUGCGCUUAAUUGUGCAGCGCAUUCCGUAUACGCCGUGUGCGACGAACCCUGAAUAGA---------GCCUGCCUAAUGGAGCAA ....(((....)))((((.((((.((((.((....(((((((...))))))).......((....)).---------)))))).)))).)))).. ( -23.10) >DroSec_CAF1 27328 95 - 1 AGCCGCACAAAUGCGCUUAAUUGUGCAGCGCAUUGCGUAUGCGGCGUGGGUGCAGCUCGUUGCGUAUACGCCGCGUGUGCUGCCUAAUGGAGCGG ..((((...((((((((.........))))))))((((((((((((((..(((((....)))))..))))))))))))))..((....)).)))) ( -45.90) >DroSim_CAF1 28621 95 - 1 AGCCGCACAAAAGCGCUUAAUUGUGCAGCGCAUUGCGUAUGCGGCGUGGGUGCAGCUCGUUGCGUAUACGCCGCGUGUGCUGCCUAAUGGAGCGA .............(((((.((((.(((((((((.......((((((((..(((((....)))))..))))))))))))))))).)))).))))). ( -44.61) >DroYak_CAF1 28640 95 - 1 AGCCGCACAAAUGCGCUUAAUUGUGCAGCGCAUUGCGUAUGCGGCGUGGGUGCAGCUCGUUGCGUAUACGCCGCGUGUGCUGCCUAAUGGAGCGA ....(((....)))((((.((((.(((((((((.......((((((((..(((((....)))))..))))))))))))))))).)))).)))).. ( -44.91) >DroPer_CAF1 27485 84 - 1 AGCCGCACAAAUGCGCUUAAUUGUGCAGUGCAUUCCGUAUACGCCGUGUGCGACGAACCCC--AUAGA---------GCCUGCCUAAUGGAGCAA .((.((((...(((((......)))))))))....(((((((...))))))).......((--((((.---------(....))).)))).)).. ( -23.30) >consensus AGCCGCACAAAUGCGCUUAAUUGUGCAGCGCAUUGCGUAUGCGGCGUGGGUGCAGCUCGUUGCGUAUACGCCGCGUGUGCUGCCUAAUGGAGCGA ....((......))((((.((((.(((((((((.......((((((((..(((((....)))))..))))))))))))))))).)))).)))).. (-26.42 = -31.53 + 5.11)


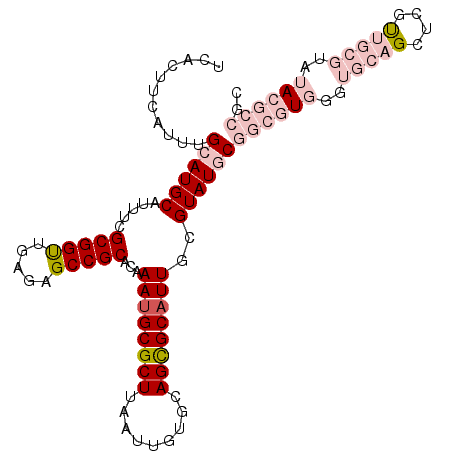
| Location | 13,430,071 – 13,430,176 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 86.93 |
| Mean single sequence MFE | -39.15 |
| Consensus MFE | -30.80 |
| Energy contribution | -33.72 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13430071 105 - 20766785 UCACUUCAUUUGCAUGCAUUUCGCGGUUGAGAGCCGCACAAAUGCGCUUAAUUGUGCAGCGCAUUGCGUAUGCGGCGUGGGUGCAGCUCGUUGCGUAUACGCCGC ...........((((((.....(((((.....)))))...((((((((.........))))))))..))))))((((((..(((((....)))))..)))))).. ( -44.20) >DroPse_CAF1 27408 99 - 1 UCGCUUCAUUUGCAUGCAUUACGCGGCUGAGAGCCGCACAAAUGCGCUUAAUUGUGCAGCGCAUUCCGUAUACGCCGUGUGCGACGAACCCUGAAUAGA------ ....(((..((((((((.....(((((.....)))))((.((((((((.........))))))))..)).......)))))))).)))...........------ ( -29.50) >DroSim_CAF1 28642 105 - 1 UCACUUCAUUUGCAUGCAUUUCGCGGUUGAGAGCCGCACAAAAGCGCUUAAUUGUGCAGCGCAUUGCGUAUGCGGCGUGGGUGCAGCUCGUUGCGUAUACGCCGC ...........((((((.....(((((.....)))))......((((......))))...))).)))....((((((((..(((((....)))))..)))))))) ( -42.40) >DroEre_CAF1 28980 105 - 1 UCACUUCAUUUGCAUGCAUUUCGCGGUUGAGAGCCGCACAAAUGCGCUUAAUUGUGCAGCGCAUUGCGUAUGCGGCGUGGGUGCAGCUCGUUGCGUAUACGCCGC ...........((((((.....(((((.....)))))...((((((((.........))))))))..))))))((((((..(((((....)))))..)))))).. ( -44.20) >DroYak_CAF1 28661 105 - 1 UCACUUCAUUUGCAUGCAUUUCGCGGUUGAGAGCCGCACAAAUGCGCUUAAUUGUGCAGCGCAUUGCGUAUGCGGCGUGGGUGCAGCUCGUUGCGUAUACGCCGC ...........((((((.....(((((.....)))))...((((((((.........))))))))..))))))((((((..(((((....)))))..)))))).. ( -44.20) >DroPer_CAF1 27503 97 - 1 UCGCUUCAUUUGCAUGCAUUACGCGGCUGAGAGCCGCACAAAUGCGCUUAAUUGUGCAGUGCAUUCCGUAUACGCCGUGUGCGACGAACCCC--AUAGA------ ..((.......))(((((((..(((((.....)))))......((((......)))))))))))..(((((((...))))))).........--.....------ ( -30.40) >consensus UCACUUCAUUUGCAUGCAUUUCGCGGUUGAGAGCCGCACAAAUGCGCUUAAUUGUGCAGCGCAUUGCGUAUGCGGCGUGGGUGCAGCUCGUUGCGUAUACGCCGC ...........((((((.....(((((.....)))))...((((((((.........))))))))..))))))((((((..(((((....)))))..)))))).. (-30.80 = -33.72 + 2.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:58 2006