
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,399,231 – 13,399,335 |
| Length | 104 |
| Max. P | 0.631810 |

| Location | 13,399,231 – 13,399,335 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.13 |
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -9.35 |
| Energy contribution | -9.68 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
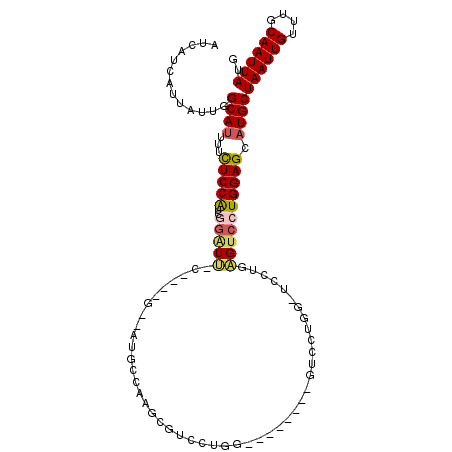
>2R_DroMel_CAF1 13399231 104 - 20766785 AUCAUCAUUAUUGGCAUUUUCUCCAUCGGAUU-C----G--AUGCCAUGCGUCCUGGAGUCCUGAGUCCUGAGUCCUUAGUCCUGGAGCAUGCUAAUUGUUUGCAAUUAUG .........((((((((...((((...(((((-(----(--((((...)))))...))))))..((..((((....))))..)))))).)))))))).............. ( -28.00) >DroSec_CAF1 16792 96 - 1 AUCAUAAUUAUUGGCAUUUUUUCCAUCGGAUU-C----G--AUGCCAAGCGUCCUGG--------GUCCUGUGUCCUGAGUCCUGGAGCAUGCUAAUUGUUUGCAAUUAUG ..(((((((((((((((...(((((..(((((-(----(--((((...)))))..((--------(........)))))))))))))).)))))))).......))))))) ( -27.51) >DroSim_CAF1 17620 96 - 1 AUCAUCAUUAUUGGCAUUUUCUCCAUCGGAUU-C----G--AUGCCAAGCGUCCUGG--------GUCCUGGAUCCUGAGUCCUGGAGCAUGCUAAUUGUUUGCAAUUAUG .........((((((((...(((((..(((((-(----(--((((...)))))..((--------(((...))))).))))))))))).)))))))).............. ( -30.00) >DroEre_CAF1 17084 82 - 1 AUCAUCAUUAUUGGCAUUUUCUCCGCUGGGUU-C----G--AUGCCAGG-------A--------GUCCUGG-------GUCCUGGAGCAUGCUAAUUGUUUGCAAUUAUG ..((((....(..((.........))..)...-.----)--)))(((((-------(--------.(....)-------.)))))).(((.((.....)).)))....... ( -21.80) >DroWil_CAF1 32226 85 - 1 --------UAUUUGCAGCUCCUCCACCGUUUG-U----GGCAUCCAUUGCUCCAUGG--------CUCCU-----CUCUGCCAUGGAGCAUGCUAAUUGUUUGCAAUUAUG --------.....(((......((((.....)-)----)).......((((((((((--------(....-----....))))))))))))))((((((....)))))).. ( -31.10) >DroYak_CAF1 16892 101 - 1 AUCAUCAUUAUUGGCAUUUUCUCCAUCAGGUUUCGAUGG--AUGCCAAGCGUGCUGA--------GUCCUGGUAGCUGAGUCCUGGAGCAUGCUAAUUGUUUGCAAUUAUG ..........((((((.....((((((.......)))))--)))))))((((((...--------.(((.((.........)).)))))))))((((((....)))))).. ( -30.10) >consensus AUCAUCAUUAUUGGCAUUUUCUCCAUCGGAUU_C____G__AUGCCAAGCGUCCUGG________GUCCUGG_UCCUGAGUCCUGGAGCAUGCUAAUUGUUUGCAAUUAUG .............((((...(((((..(((((..............................................)))))))))).))))((((((....)))))).. ( -9.35 = -9.68 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:50 2006