| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,416,055 – 2,416,166 |
| Length | 111 |
| Max. P | 0.975770 |

| Location | 2,416,055 – 2,416,166 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.12 |
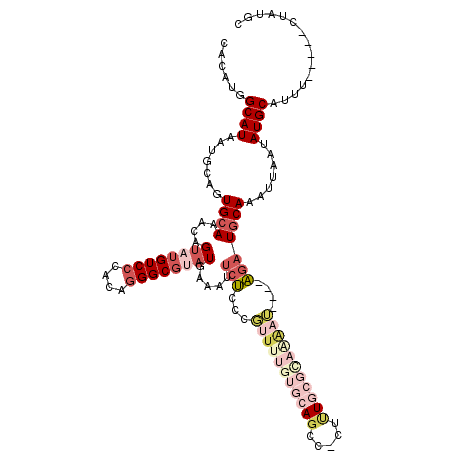
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -16.26 |
| Energy contribution | -19.66 |
| Covariance contribution | 3.39 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
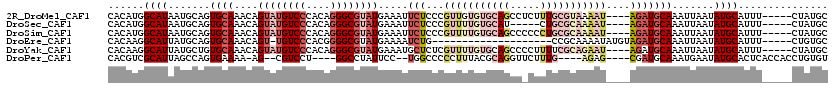
>2R_DroMel_CAF1 2416055 111 + 20766785 CACAUGGCAUAAUGCAGUGCAAACAGUAUGUCCCACAGGGCGUAUGAAAUUCUCCCGUUGUGUGCAGCCUCUUUGCGUAAAAU----AGAUGCAAAUUAAUAUGCAUUU-----CUAUGC ..(((((...((((((.((((.(((((((((((....))))))))((.....))....))).)))).....(((((((.....----..)))))))......)))))).-----))))). ( -28.50) >DroSec_CAF1 72192 106 + 1 CACAUGGCAUAAUGCAGUGCAAACAGUAUGUCCCACAGGGCGUAUGAAAUUCUCCCGUUUUGUGCAU-----CUGCGCAAAAU----AGAUGCAAAUUAAUAUGCAUUU-----CUAUGC ..(((((...((((((.((((....((((((((....)))))))).....(((...(((((((((..-----..)))))))))----)))))))........)))))).-----))))). ( -32.10) >DroSim_CAF1 29419 111 + 1 CACAUGGCAUAAUGCAGUGCAAACAGUAUGUCCCACAGGGCGUAUGAAAUUCUCCCGUUUUGUGCAGCCCCCCUGCGCAAAAU----AGAUGCAAAUUAAUAUGCAUUU-----CUAUGC ..(((((...((((((.((((....((((((((....)))))))).....(((...(((((((((((.....)))))))))))----)))))))........)))))).-----))))). ( -35.20) >DroEre_CAF1 28239 94 + 1 CACAAGGCAUUAUGCAGUGCAAACAGU-UGUCCCACGGGGCGUAUGAAAAUCUG--------------------CCGCAAAAUAUGUAGAUGCAAAUUAAUAUGCAUUU-----CUGUGC ((((.((((((((((.((....))...-.((((....)))))))))).....))--------------------))...........(((((((........)))))))-----.)))). ( -23.90) >DroYak_CAF1 28508 111 + 1 CACAAGGCAUUAUGCUGUGCAAACAGUAUGUCCCACAGGGCGUAUGAAAUGCUCUCGUUUUGUGCAGCCCCUUUUCGCAGAAU----AGAUGCAAAUUAAUAUGCAUUU-----CUAUGC .....((...((((((((....))))))))..))...(((((((..(((((....)))))..))).))))......(((....----(((((((........)))))))-----...))) ( -33.90) >DroPer_CAF1 24195 103 + 1 CACGUCGCAUUAGCCAGUGAAAA-AG--CGUCCU----GGCCUAUUCC--UGGCCCCCUUUACGCAGGUUCUUUG----AGAG----CGAUGCAAAUGAAUAUGCACUCACCACCUGUGU ............(((((.(((..-.(--(.....----.))...))))--)))).......((((((((....((----((.(----((((........)).))).))))..)))))))) ( -28.10) >consensus CACAUGGCAUAAUGCAGUGCAAACAGUAUGUCCCACAGGGCGUAUGAAAUUCUCCCGUUUUGUGCAGCC_CUUUGCGCAAAAU____AGAUGCAAAUUAAUAUGCAUUU_____CUAUGC ......((((.......((((....((((((((....)))))))).....(((...(((((((((((.....)))))))))))....))))))).......))))............... (-16.26 = -19.66 + 3.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:48 2006