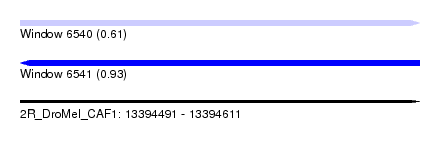
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,394,491 – 13,394,611 |
| Length | 120 |
| Max. P | 0.929231 |

| Location | 13,394,491 – 13,394,611 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.50 |
| Mean single sequence MFE | -42.78 |
| Consensus MFE | -32.77 |
| Energy contribution | -33.83 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13394491 120 + 20766785 UUGGCUCUUUAAUUGGAGCCACAAUUCUAGCGUUCUGUUCGAUGCGCCUGCGAAUGUCCGUCAGUCGCUCCUGACAGCCGCUAGCAUAGUAGCUCCAAGUGGCACGGAAAACCGAAUCCC ...(((......(((((((.((.((.((((((..(((((.(..(((.(((((......)).))).)))..).))))).)))))).)).)).)))))))..))).(((....)))...... ( -42.70) >DroGri_CAF1 15280 120 + 1 UCUUAUCCUUGAUCUGUUGCACAAAGUCAGCAUGCUGCUCGAUGCGACGACGUAUGUCCAGUUGCCGCUUCUUGCAGCCGCUGGCAAAGUAACUCCAAUAGGCCCGGAACAGUGAGUCCG ......(((......(((((.....((((((..(((((..((.(((.((((.........)))).))).))..))))).))))))...)))))......)))..((((........)))) ( -37.60) >DroSec_CAF1 12091 120 + 1 UUGGCUCUUUAAUUGGAGCCACAAUUCUAGCGUUCUGUUCGAUGCGCCUGCGAAUGUCCGUCAGUCGCUCCUGACAGCCGCUAGCAUAGUAGCUCCAAGUGGCCCGGAAAACCGAAUCCC ..((((......(((((((.((.((.((((((..(((((.(..(((.(((((......)).))).)))..).))))).)))))).)).)).)))))))..))))(((....)))...... ( -44.90) >DroSim_CAF1 12895 120 + 1 UUGGCUCUUUAAUUGGAGCCACAAUUCUAGCGUUCUGUUCGAUGCGCCUGCGAAUGUCAGUCAGUCGCUCUUGACAGCCGCUAGCAUAGUAGCUCCAAGUGGCCCGGAAAACCGAAUCCC ..((((......(((((((.((.((.((((((..(((((.((.(((.((((........).))).)))))..))))).)))))).)).)).)))))))..))))(((....)))...... ( -42.10) >DroEre_CAF1 12490 120 + 1 UUGGCUCCUUAUUUGGAGCCACAAUUCUAGCGUUCUGUUCGAUGCGCCUGCGAAUGCCCGUCAGUCGCUCCUGACAGCCGUUAGCAUAGUAGCUCCAUGUGGCCCGGAACAGCGAAUCCC ..(((..(.....((((((.((.((.((((((..(((((.(..(((.(((((......)).))).)))..).))))).)))))).)).)).)))))).)..))).(((........))). ( -41.30) >DroYak_CAF1 12516 120 + 1 UUGGCUCUUUGCUUGGAGCCACAAUUCUAGCGUUCUGUUCGAUGCGCCUGCGAAUGCCCGUCAGUCGCUCCUGACAGCCGUUAGCAUAGUAGCUCCAAGUGGCCCGGAACACCGAGUCCC ..(((((.(..((((((((.((.((.((((((..(((((.(..(((.(((((......)).))).)))..).))))).)))))).)).)).))))))))..)...((....))))))).. ( -48.10) >consensus UUGGCUCUUUAAUUGGAGCCACAAUUCUAGCGUUCUGUUCGAUGCGCCUGCGAAUGUCCGUCAGUCGCUCCUGACAGCCGCUAGCAUAGUAGCUCCAAGUGGCCCGGAAAACCGAAUCCC ..((((......(((((((.((.((.((((((..(((((.(..(((.(((((......)).))).)))..).))))).)))))).)).)).)))))))..)))).(((........))). (-32.77 = -33.83 + 1.06)

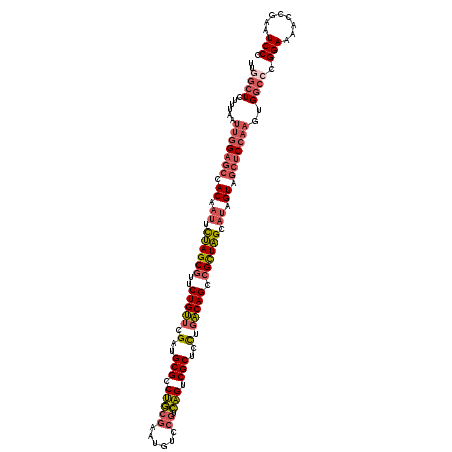
| Location | 13,394,491 – 13,394,611 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.50 |
| Mean single sequence MFE | -46.32 |
| Consensus MFE | -39.59 |
| Energy contribution | -40.48 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13394491 120 - 20766785 GGGAUUCGGUUUUCCGUGCCACUUGGAGCUACUAUGCUAGCGGCUGUCAGGAGCGACUGACGGACAUUCGCAGGCGCAUCGAACAGAACGCUAGAAUUGUGGCUCCAAUUAAAGAGCCAA .((.((((((.......)))..((((((((((....((((((.((((..(..(((.(((.(((....)))))).)))..)..))))..))))))....)))))))))).....))))).. ( -49.60) >DroGri_CAF1 15280 120 - 1 CGGACUCACUGUUCCGGGCCUAUUGGAGUUACUUUGCCAGCGGCUGCAAGAAGCGGCAACUGGACAUACGUCGUCGCAUCGAGCAGCAUGCUGACUUUGUGCAACAGAUCAAGGAUAAGA (((((.....).))))..((....)).....(((((((((..(((((.....)))))..))))......(((((.((((.((((((....))).))).)))).)).))))))))...... ( -36.70) >DroSec_CAF1 12091 120 - 1 GGGAUUCGGUUUUCCGGGCCACUUGGAGCUACUAUGCUAGCGGCUGUCAGGAGCGACUGACGGACAUUCGCAGGCGCAUCGAACAGAACGCUAGAAUUGUGGCUCCAAUUAAAGAGCCAA .((.(((((....)))))))..((((((((((....((((((.((((..(..(((.(((.(((....)))))).)))..)..))))..))))))....))))))))))............ ( -50.20) >DroSim_CAF1 12895 120 - 1 GGGAUUCGGUUUUCCGGGCCACUUGGAGCUACUAUGCUAGCGGCUGUCAAGAGCGACUGACUGACAUUCGCAGGCGCAUCGAACAGAACGCUAGAAUUGUGGCUCCAAUUAAAGAGCCAA .((.(((((....)))))))..((((((((((....((((((.((((...(((((.(((.(........)))).))).))..))))..))))))....))))))))))............ ( -46.30) >DroEre_CAF1 12490 120 - 1 GGGAUUCGCUGUUCCGGGCCACAUGGAGCUACUAUGCUAACGGCUGUCAGGAGCGACUGACGGGCAUUCGCAGGCGCAUCGAACAGAACGCUAGAAUUGUGGCUCCAAAUAAGGAGCCAA ..(((((.((((((.(.(((.(((((.....))))).....(.(((((((......))))))).).......))).)...)))))).......))))).(((((((......))))))). ( -45.20) >DroYak_CAF1 12516 120 - 1 GGGACUCGGUGUUCCGGGCCACUUGGAGCUACUAUGCUAACGGCUGUCAGGAGCGACUGACGGGCAUUCGCAGGCGCAUCGAACAGAACGCUAGAAUUGUGGCUCCAAGCAAAGAGCCAA (((((.....))))).((((.(((((((((((..(((.((.(.(((((((......))))))).).)).)))((((..((.....)).))))......)))))))))))....).))).. ( -49.90) >consensus GGGAUUCGGUGUUCCGGGCCACUUGGAGCUACUAUGCUAGCGGCUGUCAGGAGCGACUGACGGACAUUCGCAGGCGCAUCGAACAGAACGCUAGAAUUGUGGCUCCAAUUAAAGAGCCAA .((.(((((....)))))))..((((((((((....((((((.((((..((.(((.(((.((......))))).))).))..))))..))))))....))))))))))............ (-39.59 = -40.48 + 0.90)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:48 2006