| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,388,725 – 13,388,852 |
| Length | 127 |
| Max. P | 0.858160 |

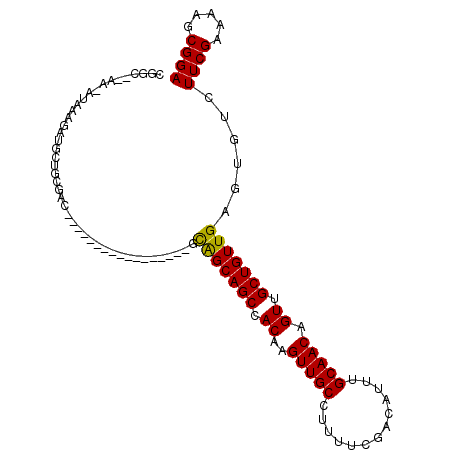
| Location | 13,388,725 – 13,388,831 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 73.49 |
| Mean single sequence MFE | -31.27 |
| Consensus MFE | -16.08 |
| Energy contribution | -15.80 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
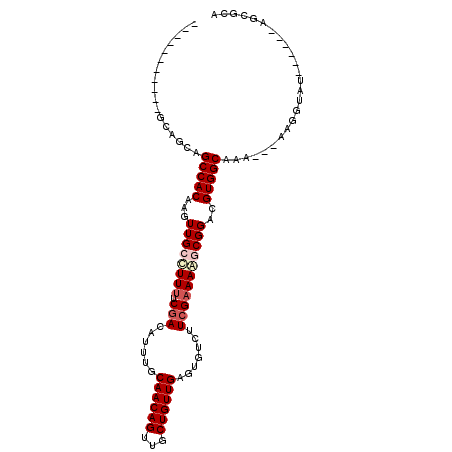
>2R_DroMel_CAF1 13388725 106 + 20766785 CGGG--AAAAAAGAGAUGCUGCGACGUCGACAGAGGCGAAAAGCAGCAGCCACAAGUUGCCUUUUCGACAUUUGCAACAGUUGCUGUUGAGUGGCUUCGAAAGUCGGA ...(--(.....(((.((((((..((((......))))....))))))(((((..((((......)))).....((((((...)))))).)))))))).....))... ( -33.50) >DroVir_CAF1 9080 88 + 1 CCAC--GA-UUUAAGAUGCUGCAAC-----------------GCAGCAGCCACAAGUUGCCUUUUCGACAUUUGCAACAGUUGCUGUUGAGUGUCUUCGAAAAGCGGA ((.(--((-(((....((((((...-----------------)))))).....)))))).(((((((((((...((((((...)))))).)))))...)))))).)). ( -26.70) >DroGri_CAF1 7831 90 + 1 CUGUGCUG-CUAAAGAUGCAGCAUC-----------------GUGGCAGCCACAAGUUGCUUUCUCGACGUUUGCAACAGUUGCUGUUGAGUGUCUUCGAAAAGCGGA .(((((((-(((..((((...))))-----------------.)))))).))))..(((((((.((((.(....((((((...)))))).....).))))))))))). ( -30.00) >DroYak_CAF1 6382 105 + 1 CGGG--AA-AAAGAGAUGCUGCGACGUCGACAGAGGCGAAAGGCAGCAGCCACAAGUUGCCUUUUCGACAUUUGCAACAGUUGCUGUUGAGUGGCUUCGAAAGUCGGA ...(--(.-.....((.((..(...(((((.((((((((..(((....))).....))))))))))))).....((((((...)))))).)..)).)).....))... ( -35.10) >DroMoj_CAF1 7269 80 + 1 CUUU--CG-CUGAAGAUG-------------------------CAGCAGCCACAAGUUGCCUUUUCGAUAUUUGCAACAGUUGCUGUUGAGUGUCUUCGAAAAGCGGA ..((--((-((((((((.-------------------------(((((((.((..(((((.............))))).)).)))))))...))))))....)))))) ( -27.32) >DroAna_CAF1 6696 101 + 1 CGGC--AA---ACGGAUGCUGCGA-GUUGGCAGACAC-AGCAGCAGCAGCCACAAGUUGCCUUUGCCACAUUUGCAACAGUUGCUGUUGAGUGACUUCGAAAGGCGGA .(((--((---..((.((((((..-((((.......)-))).)))))).)).....)))))((((((...((((....(((..((....))..))).)))).)))))) ( -35.00) >consensus CGGC__AA_AUAAAGAUGCUGCGAC_________________GCAGCAGCCACAAGUUGCCUUUUCGACAUUUGCAACAGUUGCUGUUGAGUGUCUUCGAAAAGCGGA ...........................................(((((((.((..(((((.............))))).)).)))))))......((((.....)))) (-16.08 = -15.80 + -0.28)

| Location | 13,388,725 – 13,388,831 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 73.49 |
| Mean single sequence MFE | -26.94 |
| Consensus MFE | -16.19 |
| Energy contribution | -16.90 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
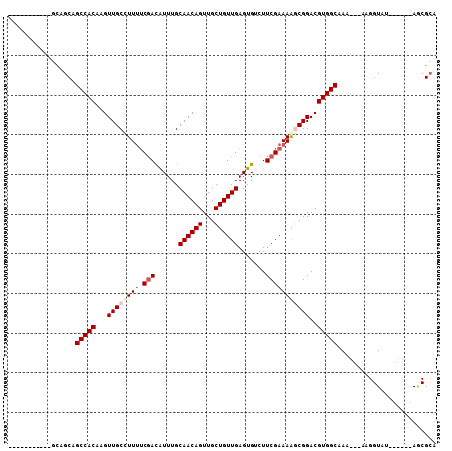
>2R_DroMel_CAF1 13388725 106 - 20766785 UCCGACUUUCGAAGCCACUCAACAGCAACUGUUGCAAAUGUCGAAAAGGCAACUUGUGGCUGCUGCUUUUCGCCUCUGUCGACGUCGCAGCAUCUCUUUUUU--CCCG ..((.....)).((((((.((((((...))))))....((((.....))))....))))))(((((...((((....).)))....)))))...........--.... ( -26.90) >DroVir_CAF1 9080 88 - 1 UCCGCUUUUCGAAGACACUCAACAGCAACUGUUGCAAAUGUCGAAAAGGCAACUUGUGGCUGCUGC-----------------GUUGCAGCAUCUUAAA-UC--GUGG .((((..(((((.......((((((...))))))......)))))(((....)))...(((((...-----------------...)))))........-..--)))) ( -25.32) >DroGri_CAF1 7831 90 - 1 UCCGCUUUUCGAAGACACUCAACAGCAACUGUUGCAAACGUCGAGAAAGCAACUUGUGGCUGCCAC-----------------GAUGCUGCAUCUUUAG-CAGCACAG ...(((((((((.......((((((...))))))......))))).))))...((((((...))))-----------------))((((((.......)-)))))... ( -28.32) >DroYak_CAF1 6382 105 - 1 UCCGACUUUCGAAGCCACUCAACAGCAACUGUUGCAAAUGUCGAAAAGGCAACUUGUGGCUGCUGCCUUUCGCCUCUGUCGACGUCGCAGCAUCUCUUU-UU--CCCG ..((.....)).((((((.((((((...))))))....((((.....))))....))))))(((((...((((....).)))....)))))........-..--.... ( -26.90) >DroMoj_CAF1 7269 80 - 1 UCCGCUUUUCGAAGACACUCAACAGCAACUGUUGCAAAUAUCGAAAAGGCAACUUGUGGCUGCUG-------------------------CAUCUUCAG-CG--AAAG ..((((....(((((....((.((((.((.(((((.............)))))..)).)))).))-------------------------..)))))))-))--.... ( -21.02) >DroAna_CAF1 6696 101 - 1 UCCGCCUUUCGAAGUCACUCAACAGCAACUGUUGCAAAUGUGGCAAAGGCAACUUGUGGCUGCUGCUGCU-GUGUCUGCCAAC-UCGCAGCAUCCGU---UU--GCCG ...((((((....(((((.((((((...)))))).....))))))))))).......(((.((.(.((((-(((.........-.))))))).).))---..--))). ( -33.20) >consensus UCCGCCUUUCGAAGACACUCAACAGCAACUGUUGCAAAUGUCGAAAAGGCAACUUGUGGCUGCUGC_________________GUCGCAGCAUCUUUAG_UU__ACCG ...(((((((((.......((((((...))))))......))).)))))).......((.((((((....................)))))).))............. (-16.19 = -16.90 + 0.71)

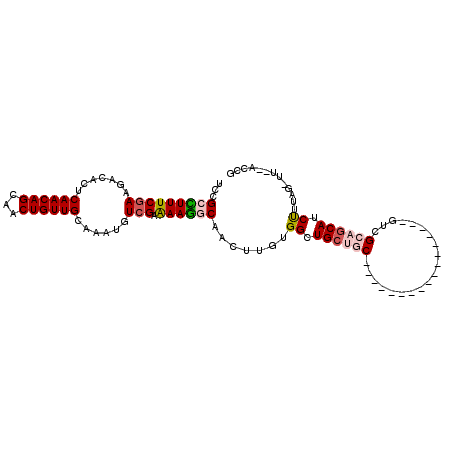
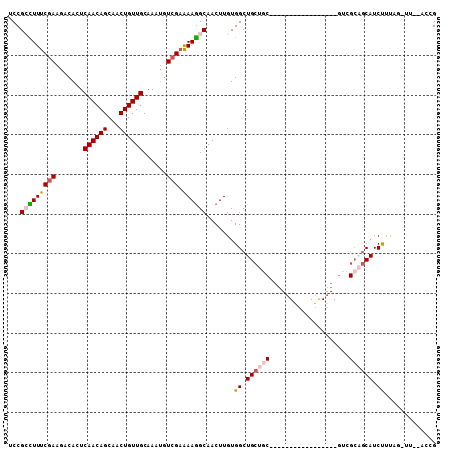
| Location | 13,388,754 – 13,388,852 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.37 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -21.22 |
| Energy contribution | -22.05 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13388754 98 + 20766785 AGAGGCGAAAAGCAGCAGCCACAAGUUGCCUUUUCGACAUUUGCAACAGUUGCUGUUGAGUGGCUUCGAAAGUCGGACGUGGCGA----AAGGUAU------AGCGCA ....(((....((....(((((.....(((..(((((((...(((.....)))))))))).)))((((.....)))).)))))..----...))..------..))). ( -30.00) >DroVir_CAF1 9102 97 + 1 -----------GCAGCAGCCACAAGUUGCCUUUUCGACAUUUGCAACAGUUGCUGUUGAGUGUCUUCGAAAAGCGGACGUGGCAAAUCAAAGGUAGCGGAACAGCGCG -----------...((.(((((...((((.((((((((((...((((((...)))))).)))))...)))))))))..)))))............((......)))). ( -30.80) >DroGri_CAF1 7855 97 + 1 -----------GUGGCAGCCACAAGUUGCUUUCUCGACGUUUGCAACAGUUGCUGUUGAGUGUCUUCGAAAAGCGGACGUGGCAAAUUAAAGGUAACGCAGCACCGCG -----------...((.(((((...(((((((.((((.(....((((((...)))))).....).)))))))))))..)))))........(((........))))). ( -30.60) >DroYak_CAF1 6410 98 + 1 AGAGGCGAAAGGCAGCAGCCACAAGUUGCCUUUUCGACAUUUGCAACAGUUGCUGUUGAGUGGCUUCGAAAGUCGGACGUGGCGA----AAGGUAU------AGCGCA ....(((.((((((((........))))))))((((((.((((....(((..((....))..))).)))).)))))).(((.(..----..).)))------..))). ( -32.20) >DroMoj_CAF1 7284 96 + 1 ------------CAGCAGCCACAAGUUGCCUUUUCGAUAUUUGCAACAGUUGCUGUUGAGUGUCUUCGAAAAGCGGACGUGGCAAAUCAAAGGUAGCGGACGAACGCG ------------.....(((((...((((.(((((((......((((((...)))))).......)))))))))))..)))))............(((......))). ( -30.72) >DroAna_CAF1 6721 98 + 1 AGACAC-AGCAGCAGCAGCCACAAGUUGCCUUUGCCACAUUUGCAACAGUUGCUGUUGAGUGACUUCGAAAGGCGGACGUGGCAAA---AAGGUAU------AGCGGA ......-.((.(((((........))))).((((((((.(((((...(((..((....))..))).(....)))))).))))))))---.......------.))... ( -32.30) >consensus ___________GCAGCAGCCACAAGUUGCCUUUUCGACAUUUGCAACAGUUGCUGUUGAGUGUCUUCGAAAAGCGGACGUGGCAAA___AAGGUAU______AGCGCA .................(((((...((((((((.(((......((((((...)))))).......)))))))))))..)))))......................... (-21.22 = -22.05 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:46 2006