| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,387,318 – 13,387,423 |
| Length | 105 |
| Max. P | 0.873689 |

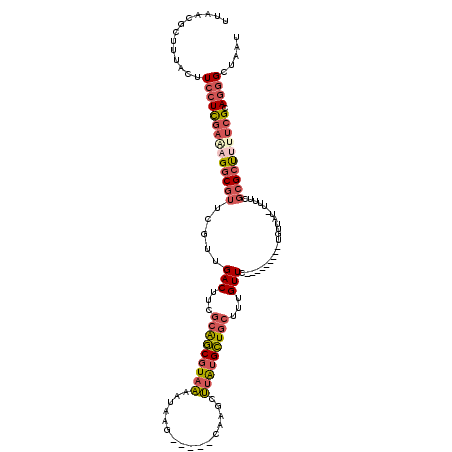
| Location | 13,387,318 – 13,387,423 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.45 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -18.64 |
| Energy contribution | -19.87 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
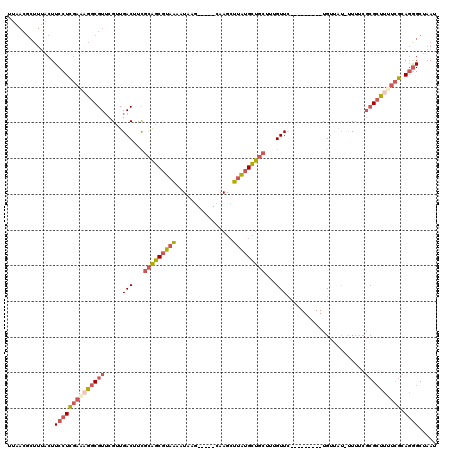
>2R_DroMel_CAF1 13387318 105 - 20766785 UUAACGCUUUACUUCCUCGAAAGGCGUUCGUUGACUUCGCAGCGUAAAAUAAG-----CAAGCUUAUGCUGCUUUGUUC---------UGUUAU-UUUUCGCGCUUUUCGCAGGGCUAAU .............((((((((((((((....((((...((((.(((..(((((-----....)))))..))).))))..---------.)))).-.....)))))))))).))))..... ( -29.80) >DroSec_CAF1 5040 106 - 1 UUAACGCUUUACUUCCUCGAAAGGCGUUCGUUGACUUUGCAGCGUAAAAUUAG-----CAAGCUUAUGCUGCUUUGUUC---------UGUUUUUUUUUCGCGCUUUUCGCAGGGCUAAU .............((((((((((((((.....(((...(((((((((......-----.....)))))))))...))).---------............)))))))))).))))..... ( -28.57) >DroSim_CAF1 5092 105 - 1 UUAACGCUUUACUUCCUCGAAAGGCGUUCGUUGACUUCGCAGCGUAAAAUAAG-----CAAGCUUAUGCUGCUUUGUUC---------UGUUAU-UUUUCGCGCUUUUCGCAGGGCUAAU .............((((((((((((((....((((...((((.(((..(((((-----....)))))..))).))))..---------.)))).-.....)))))))))).))))..... ( -29.80) >DroEre_CAF1 5098 105 - 1 UUAACGCUUUACUUCCUCGAAUGGCGUAUGUUGACUUCGCAGCGUAAAAUAAG-----CAAGCUUAUGCUGCUUUGUUU---------UGUAUU-UUUUUGCGCUUCUCGCAGAGCUAAU .....(((((.......(((..(((((((((((......)))))(((((((((-----((.((....))))).))))))---------))....-....))))))..))).))))).... ( -25.10) >DroYak_CAF1 4972 105 - 1 UUAACGCUUUACUUCCUCGACAGGCGUUCGUUGACUUCGCGGCGUAAAAUAAG-----CAAGCUUAUGCUGCUUUGUUC---------UGUUAU-UUUUCGCGCUUUUCGCAGGGCUAAU .............(((((((.((((((....((((...((((.(((..(((((-----....)))))..))).))))..---------.)))).-.....)))))).))).))))..... ( -25.50) >DroAna_CAF1 5416 106 - 1 ---------GACUUCCUUUUGCGGCGUUUGUUGACUUCUAGCCGUCUAAUAAUGGCCCCAAAGGCGAGGUGCAAUGUUUACCGGAUCCUGCAAC-CU----GGACAUUUGCAUGGCCAAU ---------.(((((((((((.(((..((((((((........))).)))))..))).)))))).)))))(((((((...((((..........-))----))))).))))......... ( -29.20) >consensus UUAACGCUUUACUUCCUCGAAAGGCGUUCGUUGACUUCGCAGCGUAAAAUAAG_____CAAGCUUAUGCUGCUUUGUUC_________UGUUAU_UUUUCGCGCUUUUCGCAGGGCUAAU .............((((((((((((((.....(((...(((((((((................)))))))))...)))......................)))))))))).))))..... (-18.64 = -19.87 + 1.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:42 2006