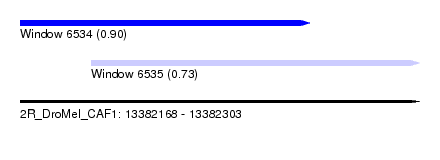
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,382,168 – 13,382,303 |
| Length | 135 |
| Max. P | 0.901110 |

| Location | 13,382,168 – 13,382,266 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.09 |
| Mean single sequence MFE | -18.67 |
| Consensus MFE | -11.91 |
| Energy contribution | -12.61 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
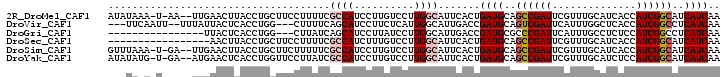
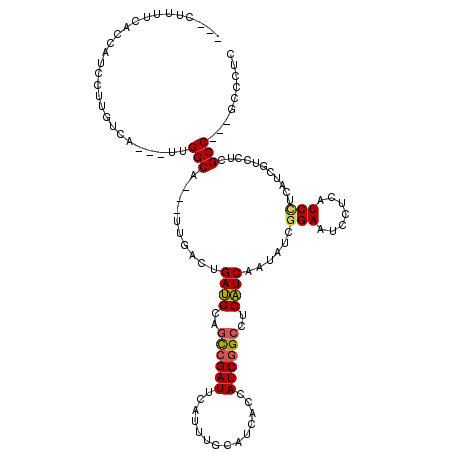
>2R_DroMel_CAF1 13382168 98 + 20766785 AUAUAAA-U-AA--UUGAACUUACCUGCUUCCUUUUCGCCAUCCUUGUCCUUGGCAUUCACUGAUGCAGCCGAUUCGUUUGCAUCACCAUCGGCAUCAUCAA .......-.-..--...........((((........((((..........))))......((((((((.((...)).)))))))).....))))....... ( -18.00) >DroVir_CAF1 1751 94 + 1 ---UUCAAUU--UUUAUUACUCACCUGG---CUUUUCAGCAUCCUUCUCAUUGGCAUUGACCGAUGCAGUCGAUUCAUUUGGCUCACCAUCGGCCUCAUCAA ---.(((((.--...............(---((....)))..((........)).)))))((((((.((((((.....))))))...))))))......... ( -17.00) >DroGri_CAF1 1 83 + 1 ----------------UUACUCACCUGG---CUUAUCAGCAUCCUUAUCCUUGGCAUUGACCGAUGCGCCCGAUUCAUUUGCCUCUCCAUCGGCCUCAUCAA ----------------........(.((---(.((((.(((.((........))...)).).)))).))).)........(((........)))........ ( -12.70) >DroSec_CAF1 1 85 + 1 -----------------AACUUACCUGCUUCCUUUUCGCCAUCUUUGUCCUUGGCAUUCACUGAUGCAGCCGAUUCGUUUGCAUCACCAUCGGCAUCAUCAA -----------------........((((........((((..........))))......((((((((.((...)).)))))))).....))))....... ( -18.00) >DroSim_CAF1 8 98 + 1 GUUUAAA-U-GA--UUGAACUUACCUGCUUCUUUUUCGCCAUCCUUGUCCUUGGCAUUCACUGAUGCAGCCGAUUCGUUUGCAUCACCAUCGGCAUCAUCAA ((((((.-.-..--)))))).....((((........((((..........))))......((((((((.((...)).)))))))).....))))....... ( -20.20) >DroYak_CAF1 8 98 + 1 AUAUAUG-U-GA--AUGAACUCACCUGGUUCCUUAUCGCCAUCCUUGUCCUUGGCAUUCACUGAUGCAGCCGAUUCGUUUGCAUCUCCAUCGGCAUCAUCAA ......(-(-((--(((((((.....)))))......((((..........))))))))))(((((..((((((..............))))))..))))). ( -26.14) >consensus ___UA_A_U_____UUGAACUCACCUGCUUCCUUUUCGCCAUCCUUGUCCUUGGCAUUCACUGAUGCAGCCGAUUCGUUUGCAUCACCAUCGGCAUCAUCAA .....................................((((..........)))).......((((..((((((..............))))))..)))).. (-11.91 = -12.61 + 0.69)

| Location | 13,382,192 – 13,382,303 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.25 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -14.25 |
| Energy contribution | -14.14 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
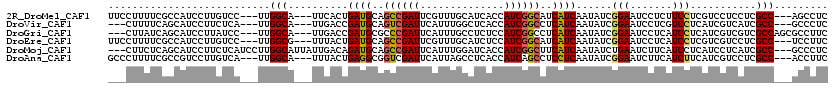
>2R_DroMel_CAF1 13382192 111 + 20766785 UUCCUUUUCGCCAUCCUUGUCC---UUGGCA---UUCACUGAUGCAGCCGAUUCGUUUGCAUCACCAUCGGCAUCAUCAAUAUCGGAAUCCUCUUCCUCGUCCUCCUCGCC---AGCCUC ......................---(((((.---.....(((((..((((((..((.......)).))))))..))))).....((((.....))))...........)))---)).... ( -22.30) >DroVir_CAF1 1774 108 + 1 ---CUUUUCAGCAUCCUUCUCA---UUGGCA---UUGACCGAUGCAGUCGAUUCAUUUGGCUCACCAUCGGCCUCAUCAAUAUCGGAAUCCUCGUCCUCAUCGUCAUCGCC---GCCCUC ---...................---..(((.---.((((.((((..(((((......(((....))))))))..))))......(((.......))).....))))..)))---...... ( -22.70) >DroGri_CAF1 13 111 + 1 ---CUUAUCAGCAUCCUUAUCC---UUGGCA---UUGACCGAUGCGCCCGAUUCAUUUGCCUCUCCAUCGGCCUCAUCAAUAUCGGAAUCCUCAUCCUCAUCGUCGUCGCCAGCGCCUUC ---...................---(((((.---.((((.((((.(.(((((..............))))).).))))......(((.......))).....))))..)))))....... ( -21.94) >DroEre_CAF1 38 111 + 1 UUCCUUUUCGCCAUCCUUGUCC---UUGGCG---UUUACUGAUGCAGCCGAUUCGUUUGCAUCUCCAUCGGCAUCAUCAAUAUCGGAAUCCUCAUCCUCGUCGUCCUCGCC---UCCUUC ((((....(((((.........---.)))))---.....(((((..((((((..............))))))..))))).....)))).......................---...... ( -22.24) >DroMoj_CAF1 37 114 + 1 ---CUUCUCAGCAUCCUUCUCAUCCUUGGCAUUAUUGACAGAUGCAGCCGAUUCAUUUGGAUCACCAUCGGCUUCAUCAAUAUCUGAAUCUUCAUCCUCAUCCUCAUCGCC---GCCCUC ---........................(((..........((((.((((((......(((....))))))))).))))......(((....)))..............)))---...... ( -20.70) >DroAna_CAF1 38 111 + 1 GCCCUUUUCGCCGUCCUUGUCA---UUGGCA---UUUACUGAGGCGGUCGAUUCAUUAGCCUCACCAUCAGCCUCCUCAAUAUCGGAAUCUUCAUCUUCAUCGUCCUCGCC---ACCUUC ......................---.((((.---.....(((((.(((.(((..............))).))).))))).....(((................)))..)))---)..... ( -18.63) >consensus ___CUUUUCACCAUCCUUGUCA___UUGGCA___UUGACUGAUGCAGCCGAUUCAUUUGCAUCACCAUCGGCCUCAUCAAUAUCGGAAUCCUCAUCCUCAUCGUCCUCGCC___GCCCUC ...........................(((..........((((..((((((..............))))))..))))......(((.......)))...........)))......... (-14.25 = -14.14 + -0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:41 2006