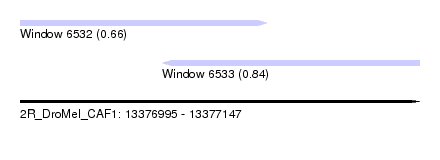
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,376,995 – 13,377,147 |
| Length | 152 |
| Max. P | 0.843388 |

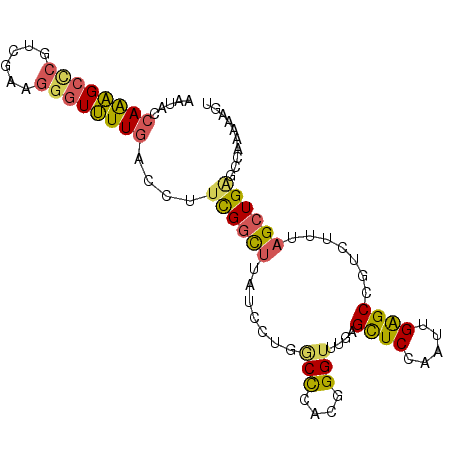
| Location | 13,376,995 – 13,377,089 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -17.89 |
| Energy contribution | -17.48 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
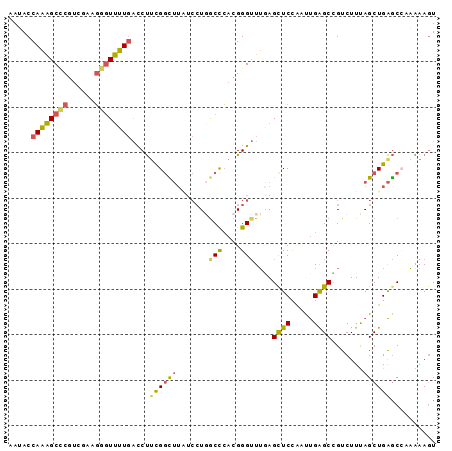
>2R_DroMel_CAF1 13376995 94 + 20766785 AAUACCAAAGCACGUCGAAGGAUUUUGACCUGUGGCUUAUCCUGACCCACGGGUUUGAGUUCUAACUGAGCCGUCUUUAGCUGAGCCAAAAAAU ..................(((((((.(((((((((.((.....)).))))))))).)))))))..((.(((........))).))......... ( -24.30) >DroPse_CAF1 75271 88 + 1 AAAGCCAGGGCUCGCCUAAUGGUCUUCGCCCUCGCU------UUGCCAACGGGAUGCAGCUCCAACCGGGCCGUCUUCAGCUGGACCAUGAAGU (((((.(((((..(((....)))....))))).)))------)).(((.(.(((.((.((((.....)))).)).))).).))).......... ( -27.70) >DroSec_CAF1 75762 94 + 1 AAUACCAAAGCCCGUCGAAGGGUUUUGACCUUCGGCUUAUCCUGGCUCACGGGUUUGAGCUCCAAUUGAGCUGUCUUUAGCUGAGCCAAAAAGU .....((((((((......))))))))......(((((.....((((((......)))))).......(((((....))))))))))....... ( -31.10) >DroSim_CAF1 79058 94 + 1 AAUACCAAAGCCCGUCGAAGGGUUUUGACCUUCGGCUUAUCCUGGCUCACGGGUUUGAGCUCUAAUUGAGCCGUCUUUAGCUGAGCCAAAAAGU .....((((((((......))))))))...(((((((.....(((((((.((((....))))....))))))).....)))))))......... ( -32.50) >DroEre_CAF1 77035 94 + 1 AAAACCAAAGCCCGUCUAAGGGUUUUGAUCUUUGGUUUUUCCUUGCCCACGGGUUUAAGCUCCGAUUGAGCUGUCUUCAGCUGAGCGAAGAAGU .....((((((((......)))))))).((((((...............(((((....)).))).((.(((((....))))).))))))))... ( -28.60) >DroYak_CAF1 78007 94 + 1 AAUACCAAAGCCCGUCUAAGGGUUUUGAUCUUUGGUUUGUCCUGGCCCACGGGUUUAAGCUCCAAUUGAGCUGUCUUUAGCUGAGCGAAGAAGU .....((((((((......)))))))).(((((((((((.((((.....))))..))))).....((.(((((....))))).))))))))... ( -30.60) >consensus AAUACCAAAGCCCGUCGAAGGGUUUUGACCUUCGGCUUAUCCUGGCCCACGGGUUUGAGCUCCAAUUGAGCCGUCUUUAGCUGAGCCAAAAAGU .....((((((((......))))))))....((((((.......(((....)))....((((.....)))).......)))))).......... (-17.89 = -17.48 + -0.41)

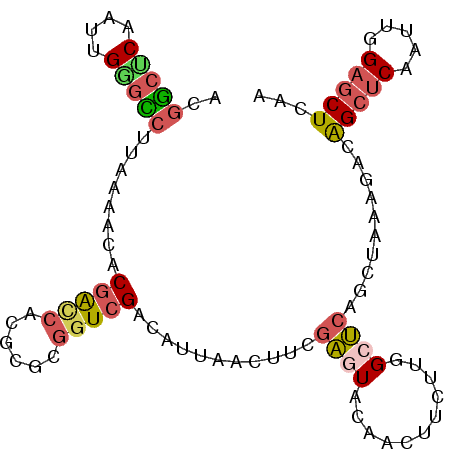
| Location | 13,377,049 – 13,377,147 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 77.48 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -15.37 |
| Energy contribution | -15.68 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13377049 98 - 20766785 ACGGCUCAAUUGGGCCUUAAAAUGCGAUCACGCGCGGUCGACAUUAACUUCGAGUACAAUUUUUUGGCUCAGCUAAAGACGGCUCAGUUAGAACUCAA ..(((((....)))))....(((((((((......))))).))))......((((.......(((((((.((((......)))).))))))))))).. ( -27.91) >DroSec_CAF1 75816 98 - 1 ACGGCUCAAUUGAGCCUUAAAACACGACCACGCGCGGUCGACAUUAACUUCGAGUACAACUUUUUGGCUCAGCUAAAGACAGCUCAAUUGGAGCUCAA ..(((((((((((((.........(((((......)))))....................((((((((...))))))))..)))))))).)))))... ( -30.10) >DroSim_CAF1 79112 98 - 1 ACGGCUCAAUUGAGCCUUAAAACACGACCACGCGCGGUCGACAUCAACUUCGAGUACAACUUUUUGGCUCAGCUAAAGACGGCUCAAUUAGAGCUCAA ..((((((((((((((........(((((......)))))....................((((((((...)))))))).))))))))).)))))... ( -33.90) >DroEre_CAF1 77089 98 - 1 ACGGCUCAAUUGGGUCUUAAAAUGCGGCCACGAGCGGUCGAUAUUAACUUUGAAUUUAACUUCUUCGCUCAGCUGAAGACAGCUCAAUCGGAGCUUAA ..(((((.((((((((.........))))..((((((..((..((((........))))..)).))))))(((((....)))))))))..)))))... ( -27.10) >DroYak_CAF1 78061 98 - 1 ACGGCACAAUUGGGCCUGAAAAUGCGACCACGAGCGGUCGAUAUUAACUUCGAGUACAACUUCUUCGCUCAGCUAAAGACAGCUCAAUUGGAGCUUAA ..(((.(((((((((.((..(((((((((......))))).))))......((((...........)))).........)))))))))))..)))... ( -27.40) >DroAna_CAF1 75799 98 - 1 GUUUGCCAAAAGGGACUACAGUCACGGCCGUCCUCAAUGGAUGUCAACUAUAGUUACAACUUCAUGGCCCAACUGAAAGCGGCGCAACUGGAGCUGAA (((.((((.(((.(((((.(((...(((.((((.....))))))).))).)))))....)))..))))..)))......((((.(.....).)))).. ( -23.00) >consensus ACGGCUCAAUUGGGCCUUAAAACACGACCACGCGCGGUCGACAUUAACUUCGAGUACAACUUCUUGGCUCAGCUAAAGACAGCUCAAUUGGAGCUCAA ..(((((....)))))........(((((......)))))...........((((...........))))..........(((((.....)))))... (-15.37 = -15.68 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:39 2006