| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,373,430 – 13,373,547 |
| Length | 117 |
| Max. P | 0.614281 |

| Location | 13,373,430 – 13,373,547 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.13 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -19.60 |
| Energy contribution | -19.96 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
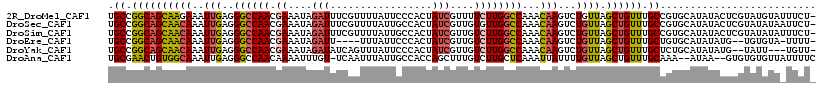
>2R_DroMel_CAF1 13373430 117 + 20766785 UGCCGGCAGCAAGAAAUUGAGGGCCAACGAAAUAGAUUUCGUUUUAUUCCCACUAUCGUUUUCUUGGCCAAACAAGUCUGUUAGCUGUUUGCCGUGCAUAUACUCGUAUGUAUUUCU- ((((((((((((((((.((((((..(((((((....))))))).....)))....))).)))))))((..((((....)))).))...)))))).))).((((......))))....- ( -30.70) >DroSec_CAF1 72173 117 + 1 UGCCGGCAGCAACAAAUUGAGGGCCAACGAAAUAGAUUUCGUUUUAUUGCCACUAUCGUUGUGUUGGCCAAACAAGUCUGUUAGCUGUUUGCCGUGCAUAUACUCGUAUAUAAUUCU- .(((((((((((((..(((..((((((((.....(((...((......))....)))....))))))))...)))...)))).))....))))).))(((((....)))))......- ( -32.90) >DroSim_CAF1 75503 117 + 1 UGCCGGCAGCAACAAAUUGAGGGCCAACGAAAUAGAUUUCGUUUUAUUGCCACUAUCGUUGUCUUGGCCAAACAAGUCUGUUAGCUGUUUGCCGUGCAUAUACUCGUAUAUAUUUCU- .(((((((((((((..(((..((((((.((....(((...((......))....)))....))))))))...)))...)))).))....))))).))(((((....)))))......- ( -30.60) >DroEre_CAF1 73493 110 + 1 UGCCGGCAGCAACAAAUUGAGGGCCAACGAAAUAGAUU----UUUAUUCCCACUAUCGUUGUCUUGGCCAAACAAGUCUGUUAGCUGUUUGCUGUGCAUAUAUG--UGUGUA-UUUU- .((..(((((((((..(((..((((((.((....(((.----............)))....))))))))...)))...)))).)))))..)).(((((((....--))))))-)...- ( -31.72) >DroYak_CAF1 74506 112 + 1 UGCCGGCAGCAACAAAUUGAGGGCCAACGAAAUAGAUAUCAGUUUAUUCCCACUAUCGUUGUCUUGGCCAAACAAGUCUGUUAGCUGUUUGCUCUGCAUAUAUG--UAUU---UGUU- .((..(((((((((..(((..((((((.((....((((...............))))....))))))))...)))...)))).)))))..))..((((....))--))..---....- ( -28.66) >DroAna_CAF1 71372 113 + 1 UGCGAACUGUGGCAAAUUGAGGGCCAACAAAAUUUGU-UCAAUUUAUUGCCACCAGCUUUGUCUUGCUCAAAUUAUUUUGUUAGCUGUUUGCAAA--AUAA--GUGUGUGUUAUUUUC .((((((((((((((((((((...(((......))))-)))))...)))))).))).)))))...((.((..(((((((((.........)))))--))))--...)).))....... ( -23.90) >consensus UGCCGGCAGCAACAAAUUGAGGGCCAACGAAAUAGAUUUCGUUUUAUUCCCACUAUCGUUGUCUUGGCCAAACAAGUCUGUUAGCUGUUUGCCGUGCAUAUACGCGUAUGUA_UUCU_ .((.((((((((((..(((..((((((.((....(((.................)))....))))))))...)))...)))).)))))).)).......................... (-19.60 = -19.96 + 0.36)
| Location | 13,373,430 – 13,373,547 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.13 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -17.20 |
| Energy contribution | -18.96 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
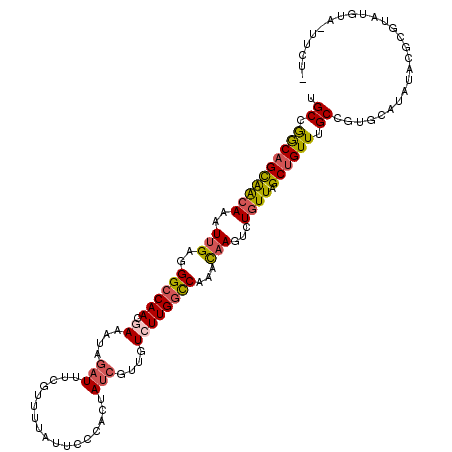
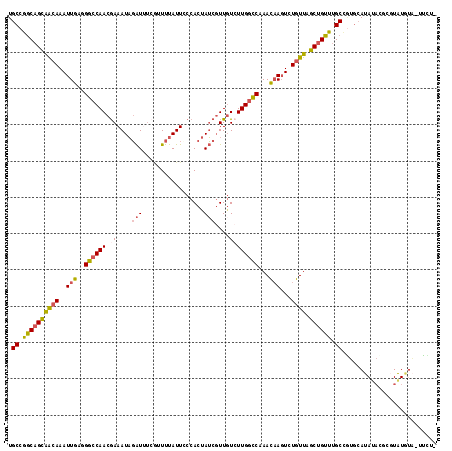
>2R_DroMel_CAF1 13373430 117 - 20766785 -AGAAAUACAUACGAGUAUAUGCACGGCAAACAGCUAACAGACUUGUUUGGCCAAGAAAACGAUAGUGGGAAUAAAACGAAAUCUAUUUCGUUGGCCCUCAAUUUCUUGCUGCCGGCA -....((((......)))).(((.(((((....((((((......).)))))(((((((((....))(((.....(((((((....)))))))..)))....))))))).)))))))) ( -31.50) >DroSec_CAF1 72173 117 - 1 -AGAAUUAUAUACGAGUAUAUGCACGGCAAACAGCUAACAGACUUGUUUGGCCAACACAACGAUAGUGGCAAUAAAACGAAAUCUAUUUCGUUGGCCCUCAAUUUGUUGCUGCCGGCA -......(((((....)))))((.(((((....((.((((((.(((...((((..(((.......))).......(((((((....)))))))))))..)))))))))))))))))). ( -34.00) >DroSim_CAF1 75503 117 - 1 -AGAAAUAUAUACGAGUAUAUGCACGGCAAACAGCUAACAGACUUGUUUGGCCAAGACAACGAUAGUGGCAAUAAAACGAAAUCUAUUUCGUUGGCCCUCAAUUUGUUGCUGCCGGCA -....((((((....))))))((.(((((....((.((((((.(((...((((..(...((....))..).....(((((((....)))))))))))..)))))))))))))))))). ( -32.70) >DroEre_CAF1 73493 110 - 1 -AAAA-UACACA--CAUAUAUGCACAGCAAACAGCUAACAGACUUGUUUGGCCAAGACAACGAUAGUGGGAAUAAA----AAUCUAUUUCGUUGGCCCUCAAUUUGUUGCUGCCGGCA -....-......--............((...((((.((((((.(((...(((((.....((((.((((((......----..)))))))))))))))..)))))))))))))...)). ( -28.10) >DroYak_CAF1 74506 112 - 1 -AACA---AAUA--CAUAUAUGCAGAGCAAACAGCUAACAGACUUGUUUGGCCAAGACAACGAUAGUGGGAAUAAACUGAUAUCUAUUUCGUUGGCCCUCAAUUUGUUGCUGCCGGCA -....---....--............((...((((.((((((.(((...(((((.....((((.((((((.((......)).)))))))))))))))..)))))))))))))...)). ( -28.50) >DroAna_CAF1 71372 113 - 1 GAAAAUAACACACAC--UUAU--UUUGCAAACAGCUAACAAAAUAAUUUGAGCAAGACAAAGCUGGUGGCAAUAAAUUGA-ACAAAUUUUGUUGGCCCUCAAUUUGCCACAGUUCGCA .(((((((.......--))))--)))((...(((((..((((....)))).(.....)..)))))(((((((...(((((-.(((......)))....)))))))))))).....)). ( -21.40) >consensus _AAAA_UACAUACGAGUAUAUGCACGGCAAACAGCUAACAGACUUGUUUGGCCAAGACAACGAUAGUGGCAAUAAAACGAAAUCUAUUUCGUUGGCCCUCAAUUUGUUGCUGCCGGCA ..........................((...((((.((((((.(((...(((((.....((((.(((((..............))))))))))))))..)))))))))))))...)). (-17.20 = -18.96 + 1.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:37 2006